For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRTAATQMPGVTREFLGLDSSTARRAGSGGHPCQGIYHRGMGRKPKVAMIAAHYQIDFSEHYLADYMATR GIGFLGWNTRYRGFESSFLLDHALVDIGVGVRWLREVQGVETVILLGNSGGGSLMAAYQANAVDKHVTPL DGMRPAAGLDDLPPADGYVASAAHPGRPDVLTAWMDASVIDENDAVATDPDLDLFNERNGPGYDAEFVAR YRAAQVARNEAITDWAEAELARVRAAGFSDRPFTVMRTWADPRMVDPALEPTKRPANMCYAGVPAKANRS THGIAAACTLRNWLGMWSLRHAQTRAEPHLARITCPALVINAEQDTGVYPSDARRIHDALGSADKTMRSV DTDHYFTTPGARSEKADTIAKWITRRW*
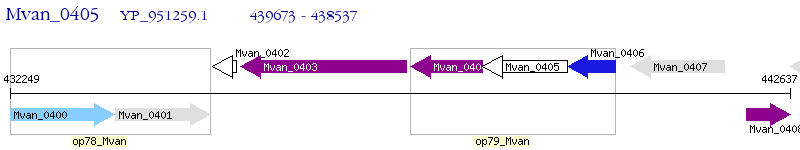
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0405 | - | - | 100% (378) | hypothetical protein Mvan_0405 |
| M. vanbaalenii PYR-1 | Mvan_1896 | - | 9e-56 | 39.62% (318) | hypothetical protein Mvan_1896 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0278c | - | 0.0 | 80.16% (378) | hypothetical protein Mb0278c |
| M. gilvum PYR-GCK | Mflv_0333 | - | 0.0 | 92.06% (378) | hypothetical protein Mflv_0333 |
| M. tuberculosis H37Rv | Rv0272c | - | 0.0 | 80.42% (378) | hypothetical protein Rv0272c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4333 | - | 1e-164 | 74.11% (367) | hypothetical protein MAB_4333 |
| M. marinum M | MMAR_0532 | - | 1e-161 | 80.66% (331) | hypothetical protein MMAR_0532 |
| M. avium 104 | MAV_4023 | - | 9e-54 | 38.68% (318) | hypothetical protein MAV_4023 |
| M. smegmatis MC2 155 | MSMEG_0605 | - | 0.0 | 83.38% (367) | hypothetical protein MSMEG_0605 |
| M. thermoresistible (build 8) | TH_0799 | - | 0.0 | 81.53% (379) | HYPOTHETICAL PROTEIN Rv0272c |
| M. ulcerans Agy99 | MUL_1196 | - | 1e-178 | 78.13% (375) | hypothetical protein MUL_1196 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0278c|M.bovis_AF2122/97 ---MTGRAATPGVIREFVGLPSRTAGRAAAGGHPCQGLYHHSVGRKPKVA
Rv0272c|M.tuberculosis_H37Rv ---MTGRAATPGVIREFVGLPSRTAGRAAAGGHPCQGLYHHSVGRKPKVA
MMAR_0532|M.marinum_M ------------------------------------------------MA
MUL_1196|M.ulcerans_Agy99 ---MSAATGTPEINREFVGLSSPTAGRAGAGGHPCQGLYHRAAGTRPTVA
Mvan_0405|M.vanbaalenii_PYR-1 MTR-TAATQMPGVTREFLGLDSSTARRAGSGGHPCQGIYHRGMGRKPKVA
Mflv_0333|M.gilvum_PYR-GCK MSR-TTAAQAPGVTREFVGLDSTTARRAGSGGHPCQGLYHRGMGRRPKVA
MSMEG_0605|M.smegmatis_MC2_155 ------------MIREFVGLESPTARRAGAGGHPCQGIYYRGVGRKPKVA
TH_0799|M.thermoresistible__bu MTRDTSASGAPGVTRQFIGVDSPTARRAGSGGHPCQGLYYRGMGRKPKVA
MAB_4333|M.abscessus_ATCC_1997 ------MPTQVTVEREFIGLPSPTAGRGGAGGHPCQGLYHRAAGTKPKVA
MAV_4023|M.avium_104 ---MKHEYDRIPYLVAFQNNSGVRDVYGGLAEITVLESYLLKPKNKPSDT
:
Mb0278c|M.bovis_AF2122/97 LIAAHYQIDFSEHY-LAEYMAIRGIGFLGWNTRFRGFESSFLLDHALVDI
Rv0272c|M.tuberculosis_H37Rv LIAAHYQIDFSEHY-LAEYMAIRGIGFLGWNTRFRGFESSFLLDHALVDI
MMAR_0532|M.marinum_M MIATHYQIDFSEHY-LADYMAARGIGFLGWNTRFRGFESNFALDHALVDI
MUL_1196|M.ulcerans_Agy99 MIATHYQIDFSEHY-LADYMAARGIGFLGWNTRFRGFESNFALDHALVDI
Mvan_0405|M.vanbaalenii_PYR-1 MIAAHYQIDFSEHY-LADYMATRGIGFLGWNTRYRGFESSFLLDHALVDI
Mflv_0333|M.gilvum_PYR-GCK MIATHYQIDFSEHY-LADYMATRGIGFLGWNTRFRGFESSFLLDHALVDI
MSMEG_0605|M.smegmatis_MC2_155 MIATHYQIDFSEHY-LADYMATRGIGFLGWNTRFRGFESSFVLDHALVDI
TH_0799|M.thermoresistible__bu VIATHYQIDFSEHY-IADYLATRGIGFLGWNTRFRGYESSFLLDHALVDI
MAB_4333|M.abscessus_ATCC_1997 FIATHYQIDFSEHY-IAEYLARHGYGFLGWNTRFRGFESQFLLDHALVDI
MAV_4023|M.avium_104 VLVFMHPIGGGAYLPMINALARAGHHVIYCNSRFRGTDSALLMEKVVEDL
.:. : *. . : : : :* * .: *:*:** :* : :::.: *:
Mb0278c|M.bovis_AF2122/97 GVGVRWLREVQGVETVVLLGNSGGGSLMAAYQSQAVDPNVTP-LDGMRPA
Rv0272c|M.tuberculosis_H37Rv GVGVRWLREVQGVETVVLLGNSGGGSLMAAYQSQAVDPNVTP-LDGMRPA
MMAR_0532|M.marinum_M GVGVRWLREVAGAETVVLLGNSGGGSLMAAYQSQAVDPNVTP-LAGTRPA
MUL_1196|M.ulcerans_Agy99 GVGVRWLREVAGAETVVLLGNSGGGSLMAAYQSQAVDPNVTP-LAGTRPA
Mvan_0405|M.vanbaalenii_PYR-1 GVGVRWLREVQGVETVILLGNSGGGSLMAAYQANAVDKHVTP-LDGMRPA
Mflv_0333|M.gilvum_PYR-GCK GVGVRWLREVQGIETVILLGNSGGGSLMAAYQANAVDKHVTP-LDGMRPA
MSMEG_0605|M.smegmatis_MC2_155 GVGVRWLREVQNIETVVLLGNSGGGSLMAAYQAQAVDPHITP-MEGMRPA
TH_0799|M.thermoresistible__bu GVGVRWLREDEGIETVILLGNSGGGSLMAAYQSQAVEPNVTP-LDGMRPA
MAB_4333|M.abscessus_ATCC_1997 GVGVRWLQEQAGVETVLLLGNSGGGSLMAAYQSQATAPKVKP-LEGMRPA
MAV_4023|M.avium_104 GECIKDAKKRLGYTKVVLAGWSGGGSLSVFYQQQAQNPTITSSPSGDGPD
* :: :: . .*:* * ****** . ** :* :.. * *
Mb0278c|M.bovis_AF2122/97 AGVTELPAADAYVAAAAHLGRPDVLTAWMDAAVIDENDPVATDPELDLFD
Rv0272c|M.tuberculosis_H37Rv AGVTELPAADAYVAAAAHPGRPDVLTAWMDAAVIDENDPVATDPELDLFD
MMAR_0532|M.marinum_M AGVNELPTADGYVATAAHPGRPEVLTAWMDAAVIDENDPVATDPDLDLFN
MUL_1196|M.ulcerans_Agy99 AGVNELPTADGYVATAAHPGRPEVLTAWMDAAVIDENDPVATDPDLDLFN
Mvan_0405|M.vanbaalenii_PYR-1 AGLDDLPPADGYVASAAHPGRPDVLTAWMDASVIDENDAVATDPDLDLFN
Mflv_0333|M.gilvum_PYR-GCK AGLDDLPAADGYVASAAHPGRPDVLTAWMDASVVDENDAVATDPDLDLFN
MSMEG_0605|M.smegmatis_MC2_155 VGINDLPRADAYVASAAHPGRPDVLTAWMDGAVLDENDPVATDPDLDLFD
TH_0799|M.thermoresistible__bu AGLNELPPADGYIASAAHPGRPEVLTAWMDAAVVDENDPVATDPELDLFN
MAB_4333|M.abscessus_ATCC_1997 EGLDGLLPAAGYVATAAHLGRPDVLTDWMDASVIDESDPASTDPALDLFN
MAV_4023|M.avium_104 LTKLDLIPADGIMLLAAHISRHGTLTEWLDASILDESDPTKRDPELDLYD
* * . : *** .* .** *:*.:::**.*.. ** ***::
Mb0278c|M.bovis_AF2122/97 ERN--GPPYSPEFISRYRSAQVKRNHTITDWAESELKRVRAAGFSDR--P
Rv0272c|M.tuberculosis_H37Rv ERN--GPPYSPEFISRYRSAQVKRNHTITDWAESELKRVRAAGFSDR--P
MMAR_0532|M.marinum_M ERN--GPPYSPEFLSRYRAAQAARNESITDWAEDELKRVRAAGFSDR--P
MUL_1196|M.ulcerans_Agy99 ERN--GPPYSPEFLSRYRAAQAARNESITDWAEDELKRVRAAGFSDR--P
Mvan_0405|M.vanbaalenii_PYR-1 ERN--GPGYDAEFVARYRAAQVARNEAITDWAEAELARVRAAGFSDR--P
Mflv_0333|M.gilvum_PYR-GCK ERN--GPSYDAEFVTRYRAAQVARNEAITDWAEAELARVRAAGFSDR--P
MSMEG_0605|M.smegmatis_MC2_155 ERN--GPPFSPDFVARYREAQIARNEAITDWAELELKRVQAAGFSDR--P
TH_0799|M.thermoresistible__bu PDN--GPPYPPEFVARYRAAQTARNHAITDWVERELKRVRAAGFSDR--P
MAB_4333|M.abscessus_ATCC_1997 QAN--GPAYAPEFVVKYREGQAARNHRITAWALEELARVRAAGFSDR--A
MAV_4023|M.avium_104 SDNPNQPPYSQEFLDRYRQAQIERNRRITAWVKAKLAELKAAGRPDDEFG
* * : :*: :** .* **. ** *. :* .::*** .*
Mb0278c|M.bovis_AF2122/97 FSVMRTWADPRMVDPSIEPTKRRPNQCYAGTPVKANRSAHGIAAACTLRG
Rv0272c|M.tuberculosis_H37Rv FSVMRTWADPRMVDPSIEPTKRRPNQCYAGTPVKANRSAHGIAAACTLRG
MMAR_0532|M.marinum_M FTVMRTWADPRMVDASIEPTNREPNLCYAGKPVRANRSARGIAAACTLRN
MUL_1196|M.ulcerans_Agy99 FTVMRTWADPRMVDASIEPTNREPNLCYAGKPVRANRSARGIAAACTLRN
Mvan_0405|M.vanbaalenii_PYR-1 FTVMRTWADPRMVDPALEPTKRPANMCYAGVPAKANRSTHGIAAACTLRN
Mflv_0333|M.gilvum_PYR-GCK FTVMRTWADPRMVDPLLEPTKRPPNMCYAGVPVKANRSTHGIAAACTLRN
MSMEG_0605|M.smegmatis_MC2_155 FTVMRTWADPRMVDPTIEPTRRQPNLCYAGVPVKANRSARGIAAATTLRN
TH_0799|M.thermoresistible__bu FTVMRTWADPRMVDPTLEPSRRPPNKCYVGEPARANRSARGIAAATTLCN
MAB_4333|M.abscessus_ATCC_1997 FTVHRTWADPRMVDPTLEPTKRPANLCYAGVPVKANRSTFGIGGATTLKN
MAV_4023|M.avium_104 FVVHGTMADPRWLDPTVDPNERTPGTCYLGDPRVVNMSPVGLARFCTLRG
* * * **** :*. ::*..* .. ** * * .* *. *:. ** .
Mb0278c|M.bovis_AF2122/97 WLGMWSLRVAQTRAAPHLARITCPALVLNAEADTGIFPSDAQQIYDGLAS
Rv0272c|M.tuberculosis_H37Rv WLGMWSLRVAQTRAAPHLARITCPALVLNAEADTGIFPSDAQQIYDGLAS
MMAR_0532|M.marinum_M WLGMWSLRVAQTRAAPHLARITCPALVINADADTGVFPSDAQCIYDGLAS
MUL_1196|M.ulcerans_Agy99 WLGMWSLRVAQARAAPHLARITCPALVINADADTGVFPSDAQCIYDGLAS
Mvan_0405|M.vanbaalenii_PYR-1 WLGMWSLRHAQTRAEPHLARITCPALVINAEQDTGVYPSDARRIHDALGS
Mflv_0333|M.gilvum_PYR-GCK WLGMWSLRHAQTRAERHLARIDCPALVINAEQDTGVFPSDAQRIYDALAG
MSMEG_0605|M.smegmatis_MC2_155 WLGMWSLRHAQTRAEPHLARITCPALVINADQDTGVYPSDAQRIFDALAG
TH_0799|M.thermoresistible__bu WLSMWSLRHSQTRAEPHLARITCPALVINAEQDTGVFPSDAQRIYDALGG
MAB_4333|M.abscessus_ATCC_1997 WLGMWSLSHAQTRAEPHLADVTVPALVINADGDTGVFPSDARRIYGALGA
MAV_4023|M.avium_104 WLSQWSYDDARGDALACGPDIAIPALVIGNLADDACTPSHTRRLFQAIGH
**. ** :: * . : ****:. * . **.:: :. .:.
Mb0278c|M.bovis_AF2122/97 SDKTQVSID-TDHYFTTPGAR---SEQADTIAKWIAKRWR----
Rv0272c|M.tuberculosis_H37Rv SDKTQVSID-TDHYFTTPGAR---SEQADTIAKWIAKRWR----
MMAR_0532|M.marinum_M TDKARASID-TDHYFTTPGAR---DEQADTITNWIAKRWQ----
MUL_1196|M.ulcerans_Agy99 TDKARASID-TDHYFTTPGAR---DEQADTITNWIAKRWQ----
Mvan_0405|M.vanbaalenii_PYR-1 ADKTMRSVD-TDHYFTTPGAR---SEKADTIAKWITRRW-----
Mflv_0333|M.gilvum_PYR-GCK DDKTLCAMD-TDHYFTTPGAR---SEKADTIAKWITRRW-----
MSMEG_0605|M.smegmatis_MC2_155 SDKTLCSID-TDHYFTTPGAR---SEQADTIARWIAKRWR----
TH_0799|M.thermoresistible__bu TDKTRVDID-TDHYFTTPGAR---SEQADVIARWIARRWR----
MAB_4333|M.abscessus_ATCC_1997 TDKSQATID-ADHYFQNPGAR---QEQADTIAEWASKRW-----
MAV_4023|M.avium_104 HDKEMHEIPGATHYYAGPDQRDALRSAVGIVTDWLVRHDFAEKA
** : : **: *. * . .. :: * ::