For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GAMTVRDIQVTFDCGDPGALAGFWAHVLGYVLQPPPAEFDTWEDALAAMQVPPERWNDASALIDPDGRRP GVFFQRVPEGKTAKNRVHLDVRAAEGLDGAERMAALERECTRLVALGATRLERHEAISLLQGGHIVMADP EGNEFCLD*
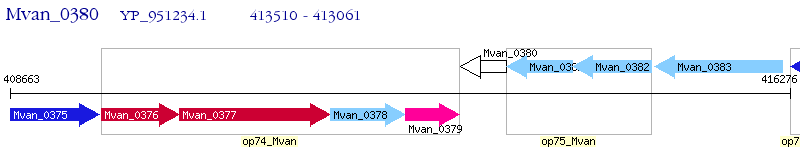
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0380 | - | - | 100% (149) | hypothetical protein Mvan_0380 |
| M. vanbaalenii PYR-1 | Mvan_5120 | - | 5e-11 | 33.56% (146) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. vanbaalenii PYR-1 | Mvan_4521 | - | 9e-11 | 29.45% (146) | hypothetical protein Mvan_4521 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0824 | - | 6e-14 | 34.93% (146) | hypothetical protein Mb0824 |
| M. gilvum PYR-GCK | Mflv_1629 | - | 4e-13 | 36.05% (147) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0801 | - | 6e-14 | 34.93% (146) | hypothetical protein Rv0801 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3166 | - | 3e-10 | 32.61% (138) | hypothetical protein MAB_3166 |
| M. marinum M | MMAR_4890 | - | 3e-16 | 39.73% (146) | hypothetical protein MMAR_4890 |
| M. avium 104 | MAV_0743 | - | 3e-14 | 36.96% (138) | glyoxalase family protein |
| M. smegmatis MC2 155 | MSMEG_5827 | - | 2e-14 | 36.69% (139) | glyoxalase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4890|M.marinum_M ----MTLSVEMVTFDCSDPARLAGWWAEHFGGTAHELLPGEFI-------
MAV_0743|M.avium_104 ----MGLNVEMVTFDCRDPARLAGWWAEQFGGTTHDILAGEFT-------
Mb0824|M.bovis_AF2122/97 ----MALKVEMVTFDCSDPAKLAGWWAEQFDGTTRELLPGEFV-------
Rv0801|M.tuberculosis_H37Rv ----MALKVEMVTFDCSDPAKLAGWWAEQFDGTTRELLPGEFV-------
Mflv_1629|M.gilvum_PYR-GCK ----MALTVEMITFDCVDPDMLASWWAGVAGGEVNAVAPGEF--------
MSMEG_5827|M.smegmatis_MC2_155 ----MGLSVEMVTVDCADPDALAQWWSQAVGGDVNVLAPGEFS-------
Mvan_0380|M.vanbaalenii_PYR-1 MGA-MTVRDIQVTFDCGDPGALAGFWAHVLG-YVLQPPPAEFDTWEDALA
MAB_3166|M.abscessus_ATCC_1997 MGLPAGLTLEAIVIDCRDAIALGRWWSEVLEWPHTIDGDGYVS-------
: :..** *. *. :*: . .
MMAR_4890|M.marinum_M -------------AVTRPAG--PRLGFQKVPDPTPGKNRVHLDFSAAD--
MAV_0743|M.avium_104 -------------AVSFPEG--LRLGFQKVSDPTPGKNRVHLDLGVVD--
Mb0824|M.bovis_AF2122/97 -------------VVARTDG--PRLGFQKVPDPAPGKNRVHLDFTTKD--
Rv0801|M.tuberculosis_H37Rv -------------VVARTDG--PRLGFQKVPDPAPGKNRVHLDFTTKD--
Mflv_1629|M.gilvum_PYR-GCK -------------VMVAAEGR-PTLGFQRVPDPTPGKNKVHLDFHAED--
MSMEG_5827|M.smegmatis_MC2_155 -------------VLVRPEG--VRLGFQKVPDPTPGKNRLHLDLSAAD--
Mvan_0380|M.vanbaalenii_PYR-1 AMQVPPERWNDASALIDPDGRRPGVFFQRVPEGKTAKNRVHLDVRAAEGL
MAB_3166|M.abscessus_ATCC_1997 --------------VFQPDRHPPLLFFMNVPDPKTAKNRLHLDFRDDD--
: . : * .*.: ..**::***. :
MMAR_4890|M.marinum_M --------VDGEVSRLAAAGATEVQRHQFGENFR--WVVMADPEGNVFCV
MAV_0743|M.avium_104 --------LDAEVSRLTAAGANEVGRHQFGESFR--WVVLADPEGNVFCL
Mb0824|M.bovis_AF2122/97 --------LDAEVLRLVAAGASEVGRHQVGESFR--WVVLADPEGNAFCV
Rv0801|M.tuberculosis_H37Rv --------LDAEVLRLVAAGASEVGRHQVGESFR--WVVLADPEGNAFCV
Mflv_1629|M.gilvum_PYR-GCK --------REAEVARLVGLGATETGRNNFGPEFE--WVVLADPEGNAFCV
MSMEG_5827|M.smegmatis_MC2_155 --------VEAEVARLVGLGAEETGRHSYGPEFS--WVVLADPEGNAFCI
Mvan_0380|M.vanbaalenii_PYR-1 DGAERMAALERECTRLVALGATRLERHEAISLLQGGHIVMADPEGNEFCL
MAB_3166|M.abscessus_ATCC_1997 --------QSALVDRFLAHGATRIDIGQG--DSP--WVVLADPEDNEFCV
. *: . ** . . :*:****.* **:
MMAR_4890|M.marinum_M VGQ------
MAV_0743|M.avium_104 VRP------
Mb0824|M.bovis_AF2122/97 AGQ------
Rv0801|M.tuberculosis_H37Rv AGQ------
Mflv_1629|M.gilvum_PYR-GCK S--------
MSMEG_5827|M.smegmatis_MC2_155 GGGD-----
Mvan_0380|M.vanbaalenii_PYR-1 D--------
MAB_3166|M.abscessus_ATCC_1997 LASRADQAK