For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGLNVEMVTFDCRDPARLAGWWAEQFGGTTHDILAGEFTAVSFPEGLRLGFQKVSDPTPGKNRVHLDLGV VDLDAEVSRLTAAGANEVGRHQFGESFRWVVLADPEGNVFCLVRP
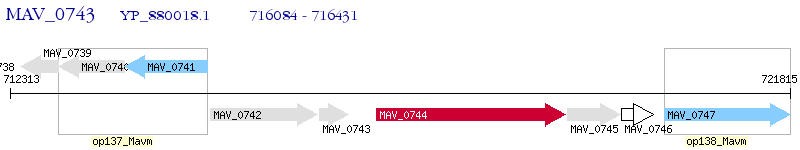
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0743 | - | - | 100% (115) | glyoxalase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0824 | - | 7e-47 | 75.00% (112) | hypothetical protein Mb0824 |
| M. gilvum PYR-GCK | Mflv_1629 | - | 6e-34 | 57.14% (112) | glyoxalase/bleomycin resistance protein/dioxygenase |
| M. tuberculosis H37Rv | Rv0801 | - | 7e-47 | 75.00% (112) | hypothetical protein Rv0801 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3166 | - | 1e-14 | 41.23% (114) | hypothetical protein MAB_3166 |
| M. marinum M | MMAR_4890 | - | 2e-49 | 76.99% (113) | hypothetical protein MMAR_4890 |
| M. smegmatis MC2 155 | MSMEG_5827 | - | 2e-39 | 61.61% (112) | glyoxalase family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5120 | - | 6e-34 | 57.14% (112) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1629|M.gilvum_PYR-GCK ----MALTVEMITFDCVDPDMLASWWAGVAGGEVNAVAPGEFVMVAAEG-
Mvan_5120|M.vanbaalenii_PYR-1 ----MSLKVEMVTFDCVDPDALADWWSKAAGGEVNAVAPGEFVMVVREG-
MSMEG_5827|M.smegmatis_MC2_155 ----MGLSVEMVTVDCADPDALAQWWSQAVGGDVNVLAPGEFSVLVRPE-
Mb0824|M.bovis_AF2122/97 ----MALKVEMVTFDCSDPAKLAGWWAEQFDGTTRELLPGEFVVVARTD-
Rv0801|M.tuberculosis_H37Rv ----MALKVEMVTFDCSDPAKLAGWWAEQFDGTTRELLPGEFVVVARTD-
MAV_0743|M.avium_104 ----MGLNVEMVTFDCRDPARLAGWWAEQFGGTTHDILAGEFTAVSFPE-
MMAR_4890|M.marinum_M ----MTLSVEMVTFDCSDPARLAGWWAEHFGGTAHELLPGEFIAVTRPA-
MAB_3166|M.abscessus_ATCC_1997 MGLPAGLTLEAIVIDCRDAIALGRWWSEVLEWP-HTIDGDGYVSVFQPDR
*.:* :..** *. *. **: . : . : :
Mflv_1629|M.gilvum_PYR-GCK -RPTLGFQRVPDPTPGKNKVHLDFHAEDREAEVARLVGLGATETGRNNFG
Mvan_5120|M.vanbaalenii_PYR-1 -GPTLGFQRVPDPTPGKNKVHLDFHAEDREAEVARLVGLGARETGRNNFG
MSMEG_5827|M.smegmatis_MC2_155 -GVRLGFQKVPDPTPGKNRLHLDLSAADVEAEVARLVGLGAEETGRHSYG
Mb0824|M.bovis_AF2122/97 -GPRLGFQKVPDPAPGKNRVHLDFTTKDLDAEVLRLVAAGASEVGRHQVG
Rv0801|M.tuberculosis_H37Rv -GPRLGFQKVPDPAPGKNRVHLDFTTKDLDAEVLRLVAAGASEVGRHQVG
MAV_0743|M.avium_104 -GLRLGFQKVSDPTPGKNRVHLDLGVVDLDAEVSRLTAAGANEVGRHQFG
MMAR_4890|M.marinum_M -GPRLGFQKVPDPTPGKNRVHLDFSAADVDGEVSRLAAAGATEVQRHQFG
MAB_3166|M.abscessus_ATCC_1997 HPPLLFFMNVPDPKTAKNRLHLDFRDDDQSALVDRFLAHGATRIDIGQ-G
* * .*.** ..**::***: * .. * *: . ** . . *
Mflv_1629|M.gilvum_PYR-GCK PEFEWVVLADPEGNAFCVS--------
Mvan_5120|M.vanbaalenii_PYR-1 PEFEWAVLADPEGNAFCVA--------
MSMEG_5827|M.smegmatis_MC2_155 PEFSWVVLADPEGNAFCIGGGD-----
Mb0824|M.bovis_AF2122/97 ESFRWVVLADPEGNAFCVAGQ------
Rv0801|M.tuberculosis_H37Rv ESFRWVVLADPEGNAFCVAGQ------
MAV_0743|M.avium_104 ESFRWVVLADPEGNVFCLVRP------
MMAR_4890|M.marinum_M ENFRWVVMADPEGNVFCVVGQ------
MAB_3166|M.abscessus_ATCC_1997 DS-PWVVLADPEDNEFCVLASRADQAK
. *.*:****.* **: