For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RARLIHTSDLDDETREDARRMVIDAFDGDFADEDWEHALGGMHAVISLRGDVIGHAAVVQRRLYVGDRAL RCGYVEGLAVREDHRGKRLGHALMDGVEQVVRGAYHLGGLSTTDLGKPLYTVRGWVPWTGPTSVLALDGA TRTPEDDGSLYVLPGELPASVTLDPTAPIACDWRNGDVW*
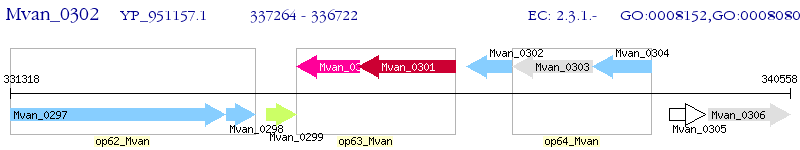
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0302 | - | - | 100% (180) | GCN5-related N-acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0268c | aac | 2e-60 | 60.67% (178) | aminoglycoside 2'-N-acetyltransferase AAC (AAC(2')-IC) |
| M. gilvum PYR-GCK | Mflv_0379 | - | 1e-82 | 76.67% (180) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv0262c | aac | 2e-60 | 60.67% (178) | aminoglycoside 2'-N-acetyltransferase AAC (AAC(2')-IC) |
| M. leprae Br4923 | MLBr_02551 | aac | 5e-59 | 57.87% (178) | aminoglycoside 2'-N-acetyltransferase |
| M. abscessus ATCC 19977 | MAB_4395 | - | 5e-53 | 57.30% (178) | aminoglycoside 2'-N-acetyltransferase |
| M. marinum M | MMAR_0522 | aac | 1e-59 | 60.67% (178) | aminoglycoside 2'-N-acetyltransferase Aac |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0434 | - | 7e-60 | 59.07% (193) | aminoglycoside 2'-N-acetyltransferase (AAC(2')-Id) |
| M. thermoresistible (build 8) | TH_0924 | - | 3e-64 | 65.17% (178) | AMINOGLYCOSIDE 2'-N-ACETYLTRANSFERASE AAC (AAC(2')-IC) |
| M. ulcerans Agy99 | MUL_1185 | aac | 5e-59 | 60.11% (178) | aminoglycoside 2'-N-acetyltransferase Aac |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0302|M.vanbaalenii_PYR-1 ---------------MRARLIHTSDLDDETREDARRMVIDAFDG------
Mflv_0379|M.gilvum_PYR-GCK ---------------MRARLVHTSDLDDETREDARRMVLDAFDG------
MSMEG_0434|M.smegmatis_MC2_155 MLTQHVSEARTRGAIHTARLIHTSDLDQETRDGARRMVIEAFRDPSGDSD
TH_0924|M.thermoresistible__bu --------------VPTARLVHTSDLDKEAFESAREMVIEAFEG------
Mb0268c|M.bovis_AF2122/97 --------MHT--QVHTARLVHTADLDSETRQDIRQMVTGAFAG------
Rv0262c|M.tuberculosis_H37Rv --------MHT--QVHTARLVHTADLDSETRQDIRQMVTGAFAG------
MMAR_0522|M.marinum_M --------MHT--EVHTARLVHTADLDSETRQGIRQMVTAAFAG------
MUL_1185|M.ulcerans_Agy99 --------MHT--EVHTARLVHTADLDSETRQGIRQMVTAAFAG------
MLBr_02551|M.leprae_Br4923 --------MDTH-HVHTARLVHTADLDGETLRRLQQMVTDAFAG------
MAB_4395|M.abscessus_ATCC_1997 MSAVSNMPTVPVIRTHTARLIHTADLDSETRCAARELLIEAFEG------
***:**:*** *: :.:: ** .
Mvan_0302|M.vanbaalenii_PYR-1 ---DFADEDWEHALGGMHAVISLRGDVIGHAAVVQRRLYVG-----DRAL
Mflv_0379|M.gilvum_PYR-GCK ---DFSEDDWEHALGGMHALICQRGDVIGHAAVVQRRLYYG-----DMAL
MSMEG_0434|M.smegmatis_MC2_155 FTDDFTDDDWDHALGGMHALISHHGALIAHGAVVQRRLMYRGPDGRGHAL
TH_0924|M.thermoresistible__bu ---EMTDTDWDNALGGMHALIQHRGTLIAHAAVVQRRLLYR-----STAL
Mb0268c|M.bovis_AF2122/97 ---DFTETDWEHTLGGMHALIWHHGAIIAHAAVIQRRLIYR-----GNAL
Rv0262c|M.tuberculosis_H37Rv ---DFTETDWEHTLGGMHALIWHHGAIIAHAAVIQRRLIYR-----GNAL
MMAR_0522|M.marinum_M ---DFTDHDWEHALGGMHALIWHRGAIIAHGAVVQRRLIYR-----GSAL
MUL_1185|M.ulcerans_Agy99 ---DFTDHDWGHALGGMHALIWHRGAIIAHGAVVQRRLIYR-----GSAL
MLBr_02551|M.leprae_Br4923 ---DFDETDWEHALGGMHALIWRHGTIIAHAAVVQRRLFYH-----GNAL
MAB_4395|M.abscessus_ATCC_1997 ---DFTEEDWEHCLGGMHALIWYHGILIAHAAVVQRRLLYQ-----DRVL
:: : ** : ******:* :* :*.*.**:**** . .*
Mvan_0302|M.vanbaalenii_PYR-1 RCGYVEGLAVREDHRGKRLGHALMDGVEQVVRGAYHLGGLSTTDLGKPLY
Mflv_0379|M.gilvum_PYR-GCK RCGYVEGVAVTEEHRGQGLGGVLMDAVEQVVRGAYHLGALGASELGEPLY
MSMEG_0434|M.smegmatis_MC2_155 RCGYVEAVAVREDRRGDGLGTAVLDALEQVIRGAYQIGALSASDIARPMY
TH_0924|M.thermoresistible__bu RCGYVEAVAVRKDWRGQGLAHALMDGIEQVLRGAYQLGALSVTEAGRHMY
Mb0268c|M.bovis_AF2122/97 RCGYVEGVAVRADWRGQRLVSALLDAVEQVMRGAYQLGALSSSARARRLY
Rv0262c|M.tuberculosis_H37Rv RCGYVEGVAVRADWRGQRLVSALLDAVEQVMRGAYQLGALSSSARARRLY
MMAR_0522|M.marinum_M RCGYVEGVAVREDWRGQGLATAVLDATEQVIRGAYELGALSSSAGARRLY
MUL_1185|M.ulcerans_Agy99 RCGYVEGVAVREDWRGQGLATAVLDATEQVIRGAYELGALSSSAGARRLY
MLBr_02551|M.leprae_Br4923 RCGYLEGVAVRKDCRGRGLVHALLDAIEQVIRGAYQFGALSSSDRARRVY
MAB_4395|M.abscessus_ATCC_1997 RTGYVEGVAVDSDWRGNGLGTAIMDAAEQVLRGAYELGALSAGDDVAPFY
* **:*.:** : ** * .::*. ***:****.:*.*. .*
Mvan_0302|M.vanbaalenii_PYR-1 TVRGWVPWTGPTSVLALDG-ATRTPEDDGSLYVLPGELPASVTLDPTAPI
Mflv_0379|M.gilvum_PYR-GCK TRRGWIRWKGPTSVLAPSG-MTPTPDDDGAVFVLPADLPPSVTLDPGAPL
MSMEG_0434|M.smegmatis_MC2_155 IARGWLSWEGPTSVLTPTEGIVRTPEDDRSLFVLPVDLPDGLELDTAREI
TH_0924|M.thermoresistible__bu TSRGWLPWLGKTSVLSPTG-IVRTPEDDRSLYVLPIS----VELDTTAEI
Mb0268c|M.bovis_AF2122/97 ASRGWLPWHGPTSVLAPTG-PVRTPDDDGTVFVLPID----ISLDTSAEL
Rv0262c|M.tuberculosis_H37Rv ASRGWLPWHGPTSVLAPTG-PVRTPDDDGTVFVLPID----ISLDTSAEL
MMAR_0522|M.marinum_M TSRGWLPWHGPTSVLSPTG-PTPTPDDDGTVFVLPVD----ASLDTSAEL
MUL_1185|M.ulcerans_Agy99 TSRGWLPWHGPTSVLSPTG-PTPTPDDDGTVFVLPVD----ASLDTSAEL
MLBr_02551|M.leprae_Br4923 MSRGWLPWLGPTSVLAPTG-VIRTPDDDGSVFVLPVG----INPDTSSGL
MAB_4395|M.abscessus_ATCC_1997 RARGWVPWLGSTPVMTADG---IVPATDEGVHVLPMS----SGVDPSGSL
***: * * *.*:: .* * :.*** *. :
Mvan_0302|M.vanbaalenii_PYR-1 ACDWRNGDVW
Mflv_0379|M.gilvum_PYR-GCK ACDWRGGDVW
MSMEG_0434|M.smegmatis_MC2_155 TCDWRSGDPW
TH_0924|M.thermoresistible__bu TCDWRDGDVW
Mb0268c|M.bovis_AF2122/97 MCDWRAGDVW
Rv0262c|M.tuberculosis_H37Rv MCDWRAGDVW
MMAR_0522|M.marinum_M MCDWRAGDVW
MUL_1185|M.ulcerans_Agy99 MCDWRAGDVW
MLBr_02551|M.leprae_Br4923 MCDWRAGNVW
MAB_4395|M.abscessus_ATCC_1997 TCDWRAGDGW
**** *: *