For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADTKGEKHTPRTEDKRGRRKTAIGYVVSDKMQKTIVVELESRKSHPLYGKIIRTTTKVKAHDENESAGIG DRVSLMETRPTSATKRWRLVEVLEKAK*
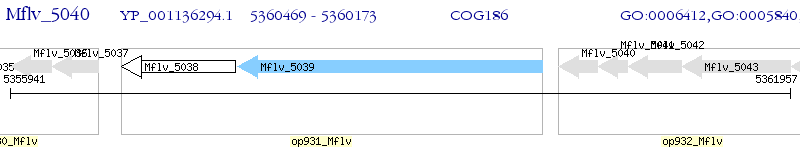
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5040 | - | - | 100% (98) | ribosomal protein S17 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0730 | rpsQ | 7e-42 | 83.51% (97) | 30S ribosomal protein S17 |
| M. tuberculosis H37Rv | Rv0710 | rpsQ | 7e-42 | 83.51% (97) | 30S ribosomal protein S17 |
| M. leprae Br4923 | MLBr_01854 | rpsQ | 3e-39 | 81.91% (94) | 30S ribosomal protein S17 |
| M. abscessus ATCC 19977 | MAB_3811c | rpsQ | 3e-42 | 87.10% (93) | 30S ribosomal protein S17 |
| M. marinum M | MMAR_1040 | rpsQ | 3e-40 | 82.98% (94) | 30S ribosomal protein S17, RpsQ |
| M. avium 104 | MAV_4462 | rpsQ | 4e-42 | 84.54% (97) | 30S ribosomal protein S17 |
| M. smegmatis MC2 155 | MSMEG_1445 | - | 7e-44 | 86.73% (98) | 30S ribosomal protein S17 |
| M. thermoresistible (build 8) | TH_0398 | rpsQ | 2e-41 | 85.11% (94) | PROBABLE 30S RIBOSOMAL PROTEIN S17 RPSQ |
| M. ulcerans Agy99 | MUL_0799 | rpsQ | 5e-39 | 80.85% (94) | 30S ribosomal protein S17 |
| M. vanbaalenii PYR-1 | Mvan_1314 | - | 1e-48 | 93.88% (98) | ribosomal protein S17 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5040|M.gilvum_PYR-GCK --MADTK-------------------------------------GEKHTP
Mvan_1314|M.vanbaalenii_PYR-1 --MADTK-------------------------------------GAKYTP
MAB_3811c|M.abscessus_ATCC_199 --MSEKKTSA------------------------------GAAPGPRHTP
MSMEG_1445|M.smegmatis_MC2_155 --MAD-------------------------------------QKGPKYTP
TH_0398|M.thermoresistible__bu --MAETKSDAQKD----------------------SAQKGSGQKGPKYTP
Mb0730|M.bovis_AF2122/97 -MMAEAKTGAKAAPRVAKAAKAAPKKAAPNDAEAIGAANAANVKGPKHTP
Rv0710|M.tuberculosis_H37Rv -MMAEAKTGAKAAPRVAKAAKAAPKKAAPNDAEAIGAANAANVKGPKHTP
MAV_4462|M.avium_104 --MAEAK-------------KAAPKKAAT------AASKDADAKGPKHTP
MMAR_1040|M.marinum_M --MAEAKTGAKATKSAAAGAAD----------------GASKEKGPKHTP
MUL_0799|M.ulcerans_Agy99 -MMAEAKTGAKATKSAAAGAAD----------------GASKEKGPKHTP
MLBr_01854|M.leprae_Br4923 MMAAEAKTSAKAAPSATTAAPKGVA----------VKGGVSKDKGPKHTP
:: * ::**
Mflv_5040|M.gilvum_PYR-GCK RTEDKRGRRKTAIGYVVSDKMQKTIVVELESRKSHPLYGKIIRTTTKVKA
Mvan_1314|M.vanbaalenii_PYR-1 RTEKPRGRRKTAIGYVVSDKMQKTIVVELESRKSHPLYGKIIRTTTKVKA
MAB_3811c|M.abscessus_ATCC_199 ATEQPRGRRKTAIGYVVSDKMQKTIVVELESRNRHSLYGKIIRTTSKVKA
MSMEG_1445|M.smegmatis_MC2_155 AAEKPRGRRKTAIGYVVSDKMQKTIVVELEDRKSHPLYGKIIRTTKKVKA
TH_0398|M.thermoresistible__bu PAEKPRGRRKTAIGYVVSDKMQKTIVVELEDRKSHPLYGKIIRTTKKVKA
Mb0730|M.bovis_AF2122/97 RTPKPRGRRKTRIGYVVSDKMQKTIVVELEDRMRHPLYGKIIRTTKKVKA
Rv0710|M.tuberculosis_H37Rv RTPKPRGRRKTRIGYVVSDKMQKTIVVELEDRMRHPLYGKIIRTTKKVKA
MAV_4462|M.avium_104 PNPKVRGRRKTRIGYVVSDKMQKTIVVELEDRVKHPLYGKIIRTTKKVKA
MMAR_1040|M.marinum_M STPKPRGRRKTRIGYVVSDKMQKTIVVELEDRVRHPLYGKIIRTTKKVKV
MUL_0799|M.ulcerans_Agy99 SPPKPSGRRKTRIGYVVSDKMQKTIVVELEDRVRHPLYGKIIRTTKKVKV
MLBr_01854|M.leprae_Br4923 SIPKVRGGRKMRIGYVVSDKMQKTIVVELEDRMRHPLYGKIIRTTKKVKA
. * ** ******************.* *.*********.***.
Mflv_5040|M.gilvum_PYR-GCK HDENESAGIGDRVSLMETRPTSATKRWRLVEVLEKAK
Mvan_1314|M.vanbaalenii_PYR-1 HDENEDAGVGDRVSLMETRPTSATKRWRLVEVLEKAK
MAB_3811c|M.abscessus_ATCC_199 HDENGDAGVGDRVSLMETRPTSATKRWRLVEVLEKAK
MSMEG_1445|M.smegmatis_MC2_155 HDENGEAGIGDRVSLMETRPLSATKRWRLVEILEKAK
TH_0398|M.thermoresistible__bu HDEHGIAGVGDRVSLMETRPLSATKRWRLVEILEKAK
Mb0730|M.bovis_AF2122/97 HDEDSVAGIGDRVSLMETRPLSATKRWRLVEILEKAK
Rv0710|M.tuberculosis_H37Rv HDEDSVAGIGDRVSLMETRPLSATKRWRLVEILEKAK
MAV_4462|M.avium_104 HDENSIAGIGDRVSLMETRPTSATKRWRLVEILEKAK
MMAR_1040|M.marinum_M HDEHSAAGIGDRVSLMETRPLSATKRWRLVEILEKAK
MUL_0799|M.ulcerans_Agy99 HDEHSAAGIGDRVSLMETRPLSATKRWRLVEILEKAK
MLBr_01854|M.leprae_Br4923 HDENSVAGIGDRVSLMETRPLSATKRWRLVEILEKAK
***. **:*********** **********:*****