For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNGAEAPPTVDVDVTTHGDFPGIAEYAQSKIGGLTRLARRPVSYARVRLTRRHDPAVERPVIAQANLDVG GRPVRAQVEAATAQEAIDTLESRLRRRLEHLHDRWDTPRGPGAAPPWRHDDKAEREIRSFGGTPREPRII RRKSYSMAPCTVDDAVAEMELLDYDFHLFAEKGSGTAAVVYRSEPTGYRIALVAPSLAGEVAPFQGLVTI SPHAVPCLREQDALERLRRLGLPFLFYVDAAQGRASVLYRRFDGDYGVLTPAPITSER*
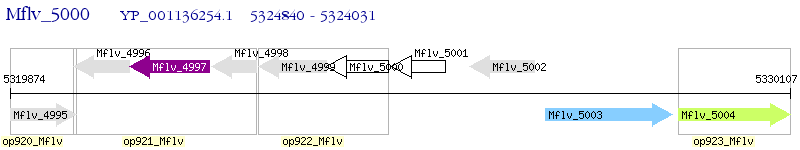
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5000 | - | - | 100% (269) | hypothetical protein Mflv_5000 |
| M. gilvum PYR-GCK | Mflv_4706 | - | 2e-08 | 31.39% (137) | sigma 54 modulation protein/ribosomal protein S30EA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0082 | - | 6e-28 | 38.50% (226) | hypothetical protein Mb0082 |
| M. tuberculosis H37Rv | Rv0079 | - | 6e-28 | 38.50% (226) | hypothetical protein Rv0079 |
| M. leprae Br4923 | MLBr_00778 | - | 1e-09 | 35.48% (124) | hypothetical protein MLBr_00778 |
| M. abscessus ATCC 19977 | MAB_3583c | - | 5e-08 | 32.09% (134) | hypothetical protein MAB_3583c |
| M. marinum M | MMAR_0944 | - | 8e-86 | 58.65% (266) | hypothetical protein MMAR_0944 |
| M. avium 104 | MAV_4204 | - | 3e-11 | 28.97% (214) | S30AE family protein |
| M. smegmatis MC2 155 | MSMEG_3935 | - | 2e-88 | 62.11% (256) | hypothetical protein MSMEG_3935 |
| M. thermoresistible (build 8) | TH_2604 | - | 6e-65 | 48.02% (252) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_0695 | - | 7e-85 | 57.99% (269) | hypothetical protein MUL_0695 |
| M. vanbaalenii PYR-1 | Mvan_1377 | - | 1e-113 | 74.90% (263) | hypothetical protein Mvan_1377 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0082|M.bovis_AF2122/97 ----------------MEPKRSRLVVCAPEPSHAREFPDVAVFSGGRANA
Rv0079|M.tuberculosis_H37Rv ----------------MEPKRSRLVVCAPEPSHAREFPDVAVFSGGRANA
Mflv_5000|M.gilvum_PYR-GCK ----------------MSNGAEAPPTVDVDVTTHGDFPGIAEYAQ-----
Mvan_1377|M.vanbaalenii_PYR-1 ----------------MSNGAEASPVVDVEVTTHGDFPGIAEYAR-----
MMAR_0944|M.marinum_M ----------------MRHKADLPPVIDVEVSARGDLPGAAEYAR-----
MUL_0695|M.ulcerans_Agy99 ----------------MRHKADLPPVIDVEVSARGDLPGAAEYAR-----
MSMEG_3935|M.smegmatis_MC2_155 --------------------------MDVDVSTDGELPGAAEYAR-----
TH_2604|M.thermoresistible__bu ----------------------VTQAFDIEISTQGTMPGAEDYVR-----
MLBr_00778|M.leprae_Br4923 MSRLTVGFGQVLNSAPVPSDEQVEPMPNVEIVFKGRNVEIPDHFR-IHVS
MAV_4204|M.avium_104 -----MDSGQILDQSSPSTDGQGQPTATAEVVFKGRNVEIPDHYR-LYVS
MAB_3583c|M.abscessus_ATCC_199 --------------------------------------------------
Mb0082|M.bovis_AF2122/97 SQAERLARAVGRVLADRGVTGGARVRLTMAN--CADGPTLVQINLQVGDT
Rv0079|M.tuberculosis_H37Rv SQAERLARAVGRVLADRGVTGGARVRLTMAN--CADGPTLVQINLQVGDT
Mflv_5000|M.gilvum_PYR-GCK SKIGGLTRLARRPVSY------ARVRLTRRHDPAVERPVIAQANLDVGGR
Mvan_1377|M.vanbaalenii_PYR-1 TKIGGLARIVHRPILR------ARVRLTRSHDPAVERPVVAQGNLDVQGR
MMAR_0944|M.marinum_M TKIGQLGRLAHRPVLS------ARVRISQHPDPAVSRPVVAQANFNVDGR
MUL_0695|M.ulcerans_Agy99 TKIGQIGRLAHRPVLS------ARVRISQHPDPAVSRPVVAQANFNVDGR
MSMEG_3935|M.smegmatis_MC2_155 EKIGRLSRRAHRPVLH------ARVRLTRHGDPAVERPVIAQANLDVNGR
TH_2604|M.thermoresistible__bu SKLGGLNKYTRRPILH------ARVRLTRHQDGSPERPVVAQANINVGGR
MLBr_00778|M.leprae_Br4923 QRLARLERFDRTIYLF------DVELDHERNRRQRKFCQRVEITARGRGP
MAV_4204|M.avium_104 QKLARLERFDRSIYLF------DVELKHAPNRRQRKSCQRVEITARGRGP
MAB_3583c|M.abscessus_ATCC_199 ----------------------------------------MEITARGRGP
: . .
Mb0082|M.bovis_AF2122/97 PLRAQAATAGIDDLRPALIR-LDRQIVRASAQWCPRPWP-----------
Rv0079|M.tuberculosis_H37Rv PLRAQAATAGIDDLRPALIR-LDRQIVRASAQWCPRPWP-----------
Mflv_5000|M.gilvum_PYR-GCK PVRAQVEAATAQEAIDTLESRLRRRLEHLHDRWDTPRG---PGAAPPWRH
Mvan_1377|M.vanbaalenii_PYR-1 PVRAQVEADTAQEAIDKLESRLRRRLEHLAERWEAPRG---PEAAPPWRN
MMAR_0944|M.marinum_M LVRAQVHGATAQEAIDRLDARLRRRLERVAEHWEARRGQQPDAGRAEWRH
MUL_0695|M.ulcerans_Agy99 LVRAQVHGATAQEAIDRLDARLRRRLERVAEHWEARRGQQPDAGRAEWRH
MSMEG_3935|M.smegmatis_MC2_155 QVRAQVEGVNAREAVDRLEARLRSRLERIAEHWEARRGGVPAEAGREWRH
TH_2604|M.thermoresistible__bu FIRAQVEGDNVREAVDRLEDRLRLQIDRITGKWEAKRE------------
MLBr_00778|M.leprae_Br4923 VVRGEACADSFYAALESAVVKLESRLRRGKDRRKVHYG------------
MAV_4204|M.avium_104 VSRAEACADSFYAAFESAVDKLENRLRRVKDRRKVHYG------------
MAB_3583c|M.abscessus_ATCC_199 VVRGEACADTFHSALEAAVAKLESRLRRGRDRRKVHYG------------
*.:. * :: : :
Mb0082|M.bovis_AF2122/97 -DRPRRR-------LTTPAEALVTRRKPVVLRRATPLQAIAAMDAMDYDV
Rv0079|M.tuberculosis_H37Rv -DRPRRR-------LTTPAEALVTRRKPVVLRRATPLQAIAAMDAMDYDV
Mflv_5000|M.gilvum_PYR-GCK DDKAEREIRSFG---GTPREPRIIRRKSYSMAPCTVDDAVAEMELLDYDF
Mvan_1377|M.vanbaalenii_PYR-1 GEEPARAPRSVT---RSTGEPRIIRRKSFAMAPCTIDDAVAEMELLDYDF
MMAR_0944|M.marinum_M ESEPRQRDTYFP---RPPEEREVIRRKSFTMAPCTVDDAATEMGLLDYDF
MUL_0695|M.ulcerans_Agy99 ESEPRQRDTYFPYFPRPPEEREVIRRKSFTMAPCTVDDAATEMGLLDYDF
MSMEG_3935|M.smegmatis_MC2_155 ESEPARRPGYFP---RPPEERRIIRRKSFSMVPCTVDEAALEMEMLDYDF
TH_2604|M.thermoresistible__bu QPRPLHRPSFFP---RAKEERQIIRRKSYTLATATLDEAVTEMELLDYDF
MLBr_00778|M.leprae_Br4923 ------------------------DKTPVSVAEATAVAVSPEKAFNTEPA
MAV_4204|M.avium_104 ------------------------DKTPVSLAAATAVP--PRAPLPSEPQ
MAB_3583c|M.abscessus_ATCC_199 ------------------------DKTPVSLAEATAVLPADAFTPPVAVN
:.. : .*
Mb0082|M.bovis_AF2122/97 HLFTDAETGEDAVVYRAGPSGLRLARQHHVFPPGWSRCRAPAGPPVPLIV
Rv0079|M.tuberculosis_H37Rv HLFTDAETGEDAVVYRAGPSGLRLARQHHVFPPGWSRCRAPAGPPVPLIV
Mflv_5000|M.gilvum_PYR-GCK HLFAEKGSGTAAVVYRSEPTGYRIALVAPSLAG------EVAPFQGLVTI
Mvan_1377|M.vanbaalenii_PYR-1 HLFTESGSETAGVVYRGGPTGYRVALVAPAVAD------EVAPYQDSVTI
MMAR_0944|M.marinum_M HLFTEQGTGMAGVLYRGGPTGYRLALVAPATAD------QLSPFELPLSI
MUL_0695|M.ulcerans_Agy99 HLFTEQGTGMAGVLYRGGPTGYRLALVAPATAD------QLSPFELPLSI
MSMEG_3935|M.smegmatis_MC2_155 HLFTEKGTGFAAVLYKGGPTGYRLVLVIPVPAD------ELSPFEKPITI
TH_2604|M.thermoresistible__bu HLFTEKGTGQDCVLYHDD-EGYRIAQITPTSPE------ELAPHTVEVSI
MLBr_00778|M.leprae_Br4923 PAHDHGGAVTD------------------------------YEPGRIVRT
MAV_4204|M.avium_104 PAADHDGVEPDTEL--------------------------DREPGRIVRT
MAB_3583c|M.abscessus_ATCC_199 GAHSHNGAEVDDG----------------------------HGPGQIVRT
. :
Mb0082|M.bovis_AF2122/97 NSRPTPVLTEAAAVDRAREHGLPFLFFTDQATGRGQLLYSRYDGNLGLIT
Rv0079|M.tuberculosis_H37Rv NSRPTPVLTEAAAVDRAREHGLPFLFFTDQATGRGQLLYSRYDGNLGLIT
Mflv_5000|M.gilvum_PYR-GCK SPHAVPCLREQDALERLRRLGLPFLFYVDAAQGRASVLYRRFDGDYGVLT
Mvan_1377|M.vanbaalenii_PYR-1 SPQPAPCLREQDALERMALLGLPFLFFIDAAQGRASLLYRRYDGAYGLIT
MMAR_0944|M.marinum_M SPHPAPILTEAEAAERLGVLGLPFLFFIDAAQGRAGVLYHRYDGHYGLIT
MUL_0695|M.ulcerans_Agy99 SPHPAPILTEAEAAERLGVLGLPFLFFIDAAQGRAGVLYHRYDGHYGLIT
MSMEG_3935|M.smegmatis_MC2_155 STHPAPCLTQRDAVERLGLLGLPFLFYIDAAEGRASVIYRRYDGHYGLIT
TH_2604|M.thermoresistible__bu SPHPAPKLTVDEATERMNMLGVPFLYFVEAESGRGNVLYHRYDGHYGLLT
MLBr_00778|M.leprae_Br4923 KEHPAKPMSVDDALYEMELVGHDFFLFLDKQSERPSVVYRRHAYDYGLIR
MAV_4204|M.avium_104 KEHPAIPMTVDDALYEMELVGHDFFLFQDKQTERPCVVYRRHAYDYGLIR
MAB_3583c|M.abscessus_ATCC_199 KEHPAKPMTVDDALYEMELVGHDFFLFHDQSSGRPSVVYRRHAYDYGLIR
. :.. : * . * *: : : * ::* *. *::
Mb0082|M.bovis_AF2122/97 PTGDGVADGLA
Rv0079|M.tuberculosis_H37Rv PTGDGVADGLA
Mflv_5000|M.gilvum_PYR-GCK PAPITSER---
Mvan_1377|M.vanbaalenii_PYR-1 PAG--------
MMAR_0944|M.marinum_M PAG--------
MUL_0695|M.ulcerans_Agy99 PAG--------
MSMEG_3935|M.smegmatis_MC2_155 PADG-------
TH_2604|M.thermoresistible__bu PADA-------
MLBr_00778|M.leprae_Br4923 LA---------
MAV_4204|M.avium_104 LA---------
MAB_3583c|M.abscessus_ATCC_199 LGE--------