For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DSGQILDQSSPSTDGQGQPTATAEVVFKGRNVEIPDHYRLYVSQKLARLERFDRSIYLFDVELKHAPNRR QRKSCQRVEITARGRGPVSRAEACADSFYAAFESAVDKLENRLRRVKDRRKVHYGDKTPVSLAAATAVPP RAPLPSEPQPAADHDGVEPDTELDREPGRIVRTKEHPAIPMTVDDALYEMELVGHDFFLFQDKQTERPCV VYRRHAYDYGLIRLA*
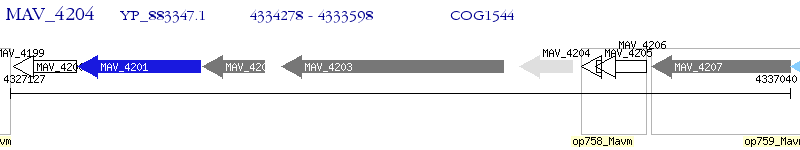
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_4204 | - | - | 100% (226) | S30AE family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3269c | - | 3e-91 | 75.55% (229) | hypothetical protein Mb3269c |
| M. gilvum PYR-GCK | Mflv_4706 | - | 7e-85 | 68.58% (226) | sigma 54 modulation protein/ribosomal protein S30EA |
| M. tuberculosis H37Rv | Rv3241c | - | 3e-91 | 75.55% (229) | hypothetical protein Rv3241c |
| M. leprae Br4923 | MLBr_00778 | - | 5e-91 | 73.68% (228) | hypothetical protein MLBr_00778 |
| M. abscessus ATCC 19977 | MAB_3583c | - | 5e-53 | 71.14% (149) | hypothetical protein MAB_3583c |
| M. marinum M | MMAR_1304 | - | 3e-91 | 72.81% (228) | hypothetical protein MMAR_1304 |
| M. smegmatis MC2 155 | MSMEG_1878 | - | 3e-84 | 70.40% (223) | S30AE family protein |
| M. thermoresistible (build 8) | TH_0995 | - | 1e-84 | 68.78% (221) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2577 | - | 2e-91 | 72.81% (228) | hypothetical protein MUL_2577 |
| M. vanbaalenii PYR-1 | Mvan_1761 | - | 9e-81 | 65.35% (228) | sigma 54 modulation protein/ribosomal protein S30EA |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1304|M.marinum_M ----------------------MTRLTVDSDQVLDQAPAQSGQPEEPQAT
MUL_2577|M.ulcerans_Agy99 ----------------------MTRLTVDSDQVLDQAPAQSGQPEEPQAT
MLBr_00778|M.leprae_Br4923 ----------------------MSRLTVGFGQVLNSAPVPSDEQVEPMPN
Mb3269c|M.bovis_AF2122/97 ---------------------------MDSGQVL----------AEPKSN
Rv3241c|M.tuberculosis_H37Rv ---------------------------MDSGQVL----------AEPKSN
MAV_4204|M.avium_104 ---------------------------MDSGQILDQSSPSTDGQGQPTAT
MAB_3583c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1878|M.smegmatis_MC2_155 --------------------MSSHSMDSSRTMVVEDDRESTAGTTPERPH
TH_0995|M.thermoresistible__bu -------------------------------MVIEDDR-PEAGRG--EPR
Mflv_4706|M.gilvum_PYR-GCK -------------MRKRIELPSMSSNSVDSDSTMVMDGEQHDAQSSDLPS
Mvan_1761|M.vanbaalenii_PYR-1 MRCLISRAREGVRSRERIELPRMSSYSVDADSTMLMD----EQSSEPTPK
MMAR_1304|M.marinum_M AEIVFKGRNVEIPDHFRTYVSQKLSRLERFDRTIYLFDVELDHERNRRQR
MUL_2577|M.ulcerans_Agy99 AEIVFKGRNVEIPDHFRTYVSQKLSRLERFDRTIYLFDVELDHERNRRQR
MLBr_00778|M.leprae_Br4923 VEIVFKGRNVEIPDHFRIHVSQRLARLERFDRTIYLFDVELDHERNRRQR
Mb3269c|M.bovis_AF2122/97 AEIVFKGRNVEIPDHFRIYVSQKLARLERFDRTIYLFDVELDHERNRRQR
Rv3241c|M.tuberculosis_H37Rv AEIVFKGRNVEIPDHFRIYVSQKLARLERFDRTIYLFDVELDHERNRRQR
MAV_4204|M.avium_104 AEVVFKGRNVEIPDHYRLYVSQKLARLERFDRSIYLFDVELKHAPNRRQR
MAB_3583c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1878|M.smegmatis_MC2_155 AEVVVKGRNVEVPDHFRTYVSEKLSRLERFDKTIYLFDVELDHERNRRQR
TH_0995|M.thermoresistible__bu AEIVFKGRNVEIPDHFRVYVSEKLSRLERFDRTIFLFDVELDHERNRRQR
Mflv_4706|M.gilvum_PYR-GCK AEVVVKGRNVEVPDHFRLYVAEKLARLERFDRTIYLFDVELDHEKNRRQR
Mvan_1761|M.vanbaalenii_PYR-1 AEVAVTGRNVEVPDHFRSYVAEKLSRLERFDRTLYLFDVELYHEKNRRQR
MMAR_1304|M.marinum_M KSCQRVEITARGRGPVVRGEACADSFYAALESAVVKVESRLRRTKDRRKV
MUL_2577|M.ulcerans_Agy99 KSCQRVEITARGRGPVVRGEACADSFYAALESAVVKVESRLRRTKDRRKV
MLBr_00778|M.leprae_Br4923 KFCQRVEITARGRGPVVRGEACADSFYAALESAVVKLESRLRRGKDRRKV
Mb3269c|M.bovis_AF2122/97 KSCQRVEITARGRGPVVRGEACADSFYAALESAVVKLESRLRRGKDRRKV
Rv3241c|M.tuberculosis_H37Rv KSCQRVEITARGRGPVVRGEACADSFYAALESAVVKLESRLRRGKDRRKV
MAV_4204|M.avium_104 KSCQRVEITARGRGPVSRAEACADSFYAAFESAVDKLENRLRRVKDRRKV
MAB_3583c|M.abscessus_ATCC_199 -----MEITARGRGPVVRGEACADTFHSALEAAVAKLESRLRRGRDRRKV
MSMEG_1878|M.smegmatis_MC2_155 KNCQHVEITARGRGPVVRGEACADSFYTAFESAVQKLEGRLRRAKDRRKI
TH_0995|M.thermoresistible__bu KNCQRVELTARGRGPVVRAEACADSFYAAMEAAVDKLEERLRRAADRRKI
Mflv_4706|M.gilvum_PYR-GCK KNCQHVEITARGRGPVVRGEACADSFYGAFEAAASKLEARLRKSKDRRKI
Mvan_1761|M.vanbaalenii_PYR-1 KNCQHVEITAKGRGPVVRGEACADSFYAALESAVSKLEGRLRKSKDRRKI
:*:**:***** *.*****:*: *:*:*. *:* ***: ****:
MMAR_1304|M.marinum_M HYGDKTPVSVAEATATVTAPEMAFAAEPAEPHEHDG----AVLDH--QPG
MUL_2577|M.ulcerans_Agy99 HYGDKTPVSVAEATATVTAPEMAFAAEPAEPHEHDG----AVLDH--QPG
MLBr_00778|M.leprae_Br4923 HYGDKTPVSVAEATAVAVSPEKAFNTEPAPAHDHGG----AVTDY--EPG
Mb3269c|M.bovis_AF2122/97 HYGDKTPVSLAEATAVVPAPENGFNTRPAEAHDHDG----AVVER--EPG
Rv3241c|M.tuberculosis_H37Rv HYGDKTPVSLAEATAVVPAPENGFNTRPAEAHDHDG----AVVER--EPG
MAV_4204|M.avium_104 HYGDKTPVSLAAATAVP--PRAPLPSEPQPAADHDGVEPDTELDR--EPG
MAB_3583c|M.abscessus_ATCC_199 HYGDKTPVSLAEATAVLPADAFTPPVAVNGAHSHNG----AEVDDGHGPG
MSMEG_1878|M.smegmatis_MC2_155 HYGDKTPVSLAEATAKDPLAGLDVSREDGEARYNDG-----VAEH--EPG
TH_0995|M.thermoresistible__bu HYGDKTPLSVAEATANMPLP--DITSPNSQSSDGAA-----TEDE--GPG
Mflv_4706|M.gilvum_PYR-GCK HYGDKTPVSLHEATALDRLDA-AFAKPTETS-----AVEAPVDDH--EPG
Mvan_1761|M.vanbaalenii_PYR-1 HYGDKTPRSLREACSPEQLEG-VFTPPAEASLSLDGQSAQPLDEH--EPG
******* *: * : . : **
MMAR_1304|M.marinum_M RVVRTKEHPAKPMSVDDALYEMELVGHDFFLFFDQETERPTVVYRRHGYD
MUL_2577|M.ulcerans_Agy99 RVVRTKEHPAKPMSVDDALYEMELVGHDFFLFFDQETERPTVVYRRHGYD
MLBr_00778|M.leprae_Br4923 RIVRTKEHPAKPMSVDDALYEMELVGHDFFLFLDKQSERPSVVYRRHAYD
Mb3269c|M.bovis_AF2122/97 RIVRTKEHPAKPMSVDDALYQMELVGHDFFLFYDKDTERPSVVYRRHAYD
Rv3241c|M.tuberculosis_H37Rv RIVRTKEHPAKPMSVDDALYQMELVGHDFFLFYDKDTERPSVVYRRHAYD
MAV_4204|M.avium_104 RIVRTKEHPAIPMTVDDALYEMELVGHDFFLFQDKQTERPCVVYRRHAYD
MAB_3583c|M.abscessus_ATCC_199 QIVRTKEHPAKPMTVDDALYEMELVGHDFFLFHDQSSGRPSVVYRRHAYD
MSMEG_1878|M.smegmatis_MC2_155 RIVRIKDHPATPMTVDDALYEMELVGHDFFLFHDKETDRPSVVYRRHAFD
TH_0995|M.thermoresistible__bu RIVRVKEHPAVPMSVDDALYEMELVGHDFFLFHDKETDKPSVVYRRHAFD
Mflv_4706|M.gilvum_PYR-GCK RIVRTKEHPATPMTVDDALYEMELVGHDFFLFHDKESDRPCVVYRRHAYD
Mvan_1761|M.vanbaalenii_PYR-1 RIVRVKEHPATPMTVDDALYEMELVGHDFFLFHDKESDRPCVVYRRHAYD
::** *:*** **:******:*********** *:.: :* ******.:*
MMAR_1304|M.marinum_M YGLIRLS-
MUL_2577|M.ulcerans_Agy99 YGLIRLS-
MLBr_00778|M.leprae_Br4923 YGLIRLA-
Mb3269c|M.bovis_AF2122/97 YGLIRLA-
Rv3241c|M.tuberculosis_H37Rv YGLIRLA-
MAV_4204|M.avium_104 YGLIRLA-
MAB_3583c|M.abscessus_ATCC_199 YGLIRLGE
MSMEG_1878|M.smegmatis_MC2_155 YGLIRLA-
TH_0995|M.thermoresistible__bu YGLIRLA-
Mflv_4706|M.gilvum_PYR-GCK YGLIKLA-
Mvan_1761|M.vanbaalenii_PYR-1 YGLIKLA-
****:*.