For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PKKIDPNVRERCVRQVLDTLSQYPSVTAACEAVARREGVGKESVRRWVVQAQIDGGQRHGATSEELAEIK ELKAKVRRLEEDNE*
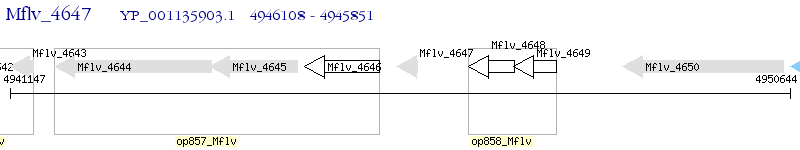
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4647 | - | - | 100% (85) | putative transposase |
| M. gilvum PYR-GCK | Mflv_4636 | - | 2e-44 | 100.00% (85) | transposase IS3/IS911 family protein |
| M. gilvum PYR-GCK | Mflv_0600 | - | 3e-09 | 41.67% (72) | Fis family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2839c | - | 2e-13 | 48.15% (81) | transposase |
| M. tuberculosis H37Rv | Rv3474 | - | 2e-13 | 48.15% (81) | transposase IS6110 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2087 | - | 1e-05 | 33.33% (54) | transposase |
| M. marinum M | MMAR_2547 | - | 5e-12 | 44.58% (83) | transposase, ISMyma03_aa1 |
| M. avium 104 | MAV_3272 | - | 9e-09 | 38.89% (72) | ISAfe7, transposase OrfA |
| M. smegmatis MC2 155 | MSMEG_1716 | - | 2e-09 | 41.67% (72) | IS3 family protein element, transposase orfA |
| M. thermoresistible (build 8) | TH_1992 | - | 6e-12 | 45.24% (84) | POSSIBLE TRANSPOSASE FOR INSERTION ELEMENT IS6110 |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5972 | - | 5e-07 | 38.89% (72) | transposase IS3/IS911 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2839c|M.bovis_AF2122/97 -----------------------------------------MSGGSSRRY
Rv3474|M.tuberculosis_H37Rv -----------------------------------------MSGGSSRRY
MMAR_2547|M.marinum_M -----------------------------------------MARAS--KY
TH_1992|M.thermoresistible__bu VLAKLIWQQHWGTWLKFGRADKLFTRLRAARLDHTVEAEIRRLAAVDXKY
Mflv_4647|M.gilvum_PYR-GCK ---------------------------------------------MPKKI
MAV_3272|M.avium_104 -----------------------------------MPKEQSPGKPTTRRY
MSMEG_1716|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5972|M.vanbaalenii_PYR-1 -----------------------------------MPKEQSVGKPTTRRY
MAB_2087|M.abscessus_ATCC_1997 ----------------------------MTSELACPKRAGWKDVSVPRPY
Mb2839c|M.bovis_AF2122/97 PPELRERAVRMVAEIRGQHD-SEWAAISEVARLLGVGCAETVRKWVRQAQ
Rv3474|M.tuberculosis_H37Rv PPELRERAVRMVAEIRGQHD-SEWAAISEVARLLGVGCAETVRKWVRQAQ
MMAR_2547|M.marinum_M PEELRERAVRMVAEIAPQYG-SQWAAICAVAGKLGVGTPETVRTWVRRAE
TH_1992|M.thermoresistible__bu SPEFRDRALRLLDTTMEDSGVSEFEAIKSVASKLGV-SQESVRRWRRKAE
Mflv_4647|M.gilvum_PYR-GCK DPNVRERCVRQVLDTLSQYP-SVTAACEAVARREGV-GKESVRRWVVQAQ
MAV_3272|M.avium_104 SAEEKAAAVRMVRTLRAELG-TEQGTVQRVARQLGY-GVESVRTWVRQVD
MSMEG_1716|M.smegmatis_MC2_155 --------MRMVRTLREELG-TEQGTVARVARQLGY-GVESVRSWVRQAD
Mvan_5972|M.vanbaalenii_PYR-1 SAEEKAAAVRMVRALRAELG-IAQGTVQRVATQLGY-GVESVRTWVRQAD
MAB_2087|M.abscessus_ATCC_1997 PREFRDDVVRVARNRDDGVT------IEQIATDFGV-HPMTLTKWMRQAD
:* :* * :: * :.:
Mb2839c|M.bovis_AF2122/97 VDAGARPGTTTEESAELKRLRRDNAELRRANAILKTASAFFAAELDRPAR
Rv3474|M.tuberculosis_H37Rv VDAGARPGTTTEESAELKRLRRDNAELRRANAILKTASAFFAAELDRPAR
MMAR_2547|M.marinum_M VDAGQRPGVSSEQSEELRRLKRENAELRRANEILKAAAIFFGAELDRPAR
TH_1992|M.thermoresistible__bu IDAGQRPGVTSTEHAEIRKLKRENAELRRANEILKSASAFFAAELDRPAT
Mflv_4647|M.gilvum_PYR-GCK IDGGQRHGATSEELAEIKELKAKVRRLEEDNE------------------
MAV_3272|M.avium_104 IDEGLAPGVTTSESKKVKELEQEIRELKRANEILKRAASFFGAELDRQHK
MSMEG_1716|M.smegmatis_MC2_155 IDDGHAPGVTTAESAKVKELEQEIRELKRANEILKRAASFFGAELDRQHK
Mvan_5972|M.vanbaalenii_PYR-1 IDEGVAPGVTTAESTRLKALEQENRELERANEILKRAASFFGAELDRQHK
MAB_2087|M.abscessus_ATCC_1997 VDEGAKPGKSTNDSADLRELRRRNRLLEQENEVLRRAAAYLSQANLPGKG
:* * * :: : :: *. *.. *
Mb2839c|M.bovis_AF2122/97 ----
Rv3474|M.tuberculosis_H37Rv ----
MMAR_2547|M.marinum_M R---
TH_1992|M.thermoresistible__bu K---
Mflv_4647|M.gilvum_PYR-GCK ----
MAV_3272|M.avium_104 K---
MSMEG_1716|M.smegmatis_MC2_155 K---
Mvan_5972|M.vanbaalenii_PYR-1 R---
MAB_2087|M.abscessus_ATCC_1997 STRS