For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTHPELLVPDPATMRHVLGHFCTGIAVITGHDGVRPLGFACQSVTSVSLEPPYVSFCPASTSATWPLIR STGKLAVNVLAREQRELCAQFATSGGDKFRGVSWSPGHNGSPVLDGTVATIEAEVEYEHGAGDHTIVVAH VTGLRAARDTEPLLYFRGAYGGLDASAP
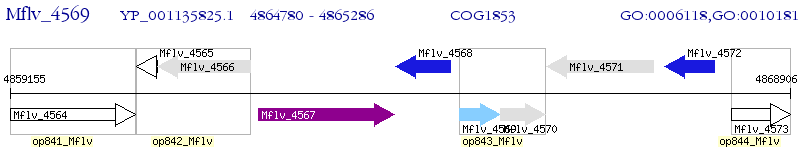
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4569 | - | - | 100% (168) | flavin reductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_1466 | - | 1e-34 | 42.95% (156) | flavin reductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0399 | - | 2e-24 | 33.54% (164) | flavin reductase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3598c | - | 1e-36 | 46.50% (157) | oxidoreductase |
| M. tuberculosis H37Rv | Rv3567c | - | 1e-36 | 46.50% (157) | oxidoreductase |
| M. leprae Br4923 | MLBr_02561 | - | 2e-21 | 34.00% (150) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0586 | - | 1e-37 | 45.96% (161) | oxidoreductase |
| M. marinum M | MMAR_5062 | - | 1e-36 | 45.86% (157) | oxidase |
| M. avium 104 | MAV_0594 | - | 6e-37 | 46.50% (157) | putative flavin reductase |
| M. smegmatis MC2 155 | MSMEG_6035 | - | 1e-37 | 47.13% (157) | nitrilotriacetate monooxygenase component B |
| M. thermoresistible (build 8) | TH_1739 | - | 5e-36 | 43.95% (157) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4138 | - | 5e-37 | 45.86% (157) | oxidase |
| M. vanbaalenii PYR-1 | Mvan_2866 | - | 1e-63 | 66.25% (160) | flavin reductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4569|M.gilvum_PYR-GCK -MTTHP------ELLVPDPATMRHVLGHFCTGIAVITGHDGVRPLGFACQ
Mvan_2866|M.vanbaalenii_PYR-1 -MTEESSAGRGSGLVTPHAPEMRRVLGHFCTGIAVVTAHDGYRPVGFTCQ
Mb3598c|M.bovis_AF2122/97 -MSAQ-----------IDPRTFRSVLGQFCTGITVITTVHDDVPVGFACQ
Rv3567c|M.tuberculosis_H37Rv -MSAQ-----------IDPRTFRSVLGQFCTGITVITTVHDDVPVGFACQ
MMAR_5062|M.marinum_M -MAADP----------IDPRTFRHVLGQFCTGITIITTVHDDVPSGFACQ
MUL_4138|M.ulcerans_Agy99 -MAADP----------IDPRTFRHVLGQFCTGITIITTVHDDVPSGFACQ
MAV_0594|M.avium_104 -MAAP-----------IDPRTFRQVLGQFCTGITIITTVHDEVPVGFACQ
MSMEG_6035|M.smegmatis_MC2_155 MTATQP----------IDPRTFRNVLGQFCTGVTVITTVHENVPVGFACQ
TH_1739|M.thermoresistible__bu -VTAEA----------IDQRTFRRVLGQFCTGVTIITTVHEGNPVGFACQ
MAB_0586|M.abscessus_ATCC_1997 -MSTPE----------IDPRTFRNVLGQFCTGITIITTTHEDVPVGFACQ
MLBr_02561|M.leprae_Br4923 -MRAMN---------SLTPSLLRDAFGHFPSGVVAIAAEVEGVREGLAAS
:* .:*:* :*:. :: *::..
Mflv_4569|M.gilvum_PYR-GCK SVTSVSLEPPYVSFCPASTSATWPLIRSTGKLAVNVLAREQRELCAQFAT
Mvan_2866|M.vanbaalenii_PYR-1 SVTSVSLDPPYISFCPANTSSSWPLIREIGAVCVNVLAADQRAVCARFAR
Mb3598c|M.bovis_AF2122/97 SFAALSLEPPLVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFGS
Rv3567c|M.tuberculosis_H37Rv SFAALSLEPPLVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFGS
MMAR_5062|M.marinum_M SFAALSLDPPLVLFCPTKVSRSWKAIEASGRFCVNVLTEKQKSVSARFGS
MUL_4138|M.ulcerans_Agy99 SFAALSLDPPLVLFCPTKVSRSWKAIEASGRFCVNVLTEKQKSVSARFGS
MAV_0594|M.avium_104 SFAALSLDPPLVLFCPTKVSRSWKAIEATGRFCVNVLTEKQKHVSARFGS
MSMEG_6035|M.smegmatis_MC2_155 SFAALSLDPPLVLFCPTKQSRAWKAIEATGRFCVNMLHENQQHVSAQFGS
TH_1739|M.thermoresistible__bu SFAALSLDPPLVLFCPTKVSRSWKAIEASGRFCVNILHEKQQHVSARFGS
MAB_0586|M.abscessus_ATCC_1997 SFAALSLEPPLVLFCPTKQSRSWAAIAASGKFCVNVLAEDQREVSARFGS
MLBr_02561|M.leprae_Br4923 TFVPVSLEPPLVSFCVQNVSTTWLKLKAVPMLGISVLGEVHDEVACTLAT
:...:**:** : ** . * :* : . :.:* : :.. :.
Mflv_4569|M.gilvum_PYR-GCK SGGDKFRGVSWSPGHNGSPVLDGTVATIEAEVEYEHGAGDHTIVVAHVTG
Mvan_2866|M.vanbaalenii_PYR-1 SGDDKFAGVDWRPGHNGAPALQGTLASIEADIEFEHAAGDHTIVVARVTG
Mb3598c|M.bovis_AF2122/97 KEPDKFAGIDWRPSELGSPIIEGSLAYIDCTVASVHDGGDHFVVFGAVES
Rv3567c|M.tuberculosis_H37Rv KEPDKFAGIDWRPSELGSPIIEGSLAYIDCTVASVHDGGDHFVVFGAVES
MMAR_5062|M.marinum_M KEPDKFAGIDWRPSELGSPIIDGSLAYIDCTVASVHDGGDHFVVFGAVHS
MUL_4138|M.ulcerans_Agy99 KESDKFAGIDWRPSELGSPIIDGSLAYIDCTVASVHDGGDHFVVFGAVHS
MAV_0594|M.avium_104 KEPDKFAGIDWHPSELGSPIIDGALAHIDCTVASVHEGGDHFVVFGAVQS
MSMEG_6035|M.smegmatis_MC2_155 KAADKFAGIDWHPSKLGSPVIDGTLAHIDCTVHSVHDGGDHFVVFGAVHS
TH_1739|M.thermoresistible__bu REPDKFAGIDWRPSDLGSPIIDGSLAHIDCTVHDVHDGGDHFVVFGKVHG
MAB_0586|M.abscessus_ATCC_1997 REPDKFAGINWRPSGLGSPIINGSLAHIDCEVANVHDGGDHWVVFGSVKE
MLBr_02561|M.leprae_Br4923 KHGDRFAGLETVSNDTGAVFIKGTSLWLESAIKQLIPAGDHTIVVLRVSQ
*:* *:. .. *: :.*: ::. : .*** :*. *
Mflv_4569|M.gilvum_PYR-GCK LRAARDT-------EPLLYFRGAYGGLDASAP------------------
Mvan_2866|M.vanbaalenii_PYR-1 LWAHEER-------SPLLFYRGGYGGFDGADHA-----------------
Mb3598c|M.bovis_AF2122/97 LSEVPAV-----KPRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLTTTTQ
Rv3567c|M.tuberculosis_H37Rv LSEVPAV-----KPRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLITTTQ
MMAR_5062|M.marinum_M LSEVPKI-----KPRPLLFYRGDYTGIEPDKTTPAQWRDDLEAFLTATTQ
MUL_4138|M.ulcerans_Agy99 LSEVPKI-----KPRPLLFYRGDYTGIEPDKTTPAQWRDDLEAFLTATTQ
MAV_0594|M.avium_104 LSEAPKI-----KPRPLLFYRGEYTGIEPDKTTPAQWRDDLEAFLTTTTQ
MSMEG_6035|M.smegmatis_MC2_155 LSEVPKT-----KPRPLLFYRGEYTGIEPDKNTPAQWRDDLEAFLTATTD
TH_1739|M.thermoresistible__bu LSEVPER-----KPRPLLFYRGEYTGIEPEKNTPAQWRDDLEAFLTATTA
MAB_0586|M.abscessus_ATCC_1997 LSERAVIPSAESAARPLLFYRGQYTGIEPDKNTPADWRNDLEAFLTATTE
MLBr_02561|M.leprae_Br4923 VTVDPDV-------APIVFHRSMFRRLGF---------------------
: *:::.*. : :
Mflv_4569|M.gilvum_PYR-GCK ----
Mvan_2866|M.vanbaalenii_PYR-1 ----
Mb3598c|M.bovis_AF2122/97 DTWL
Rv3567c|M.tuberculosis_H37Rv DTWL
MMAR_5062|M.marinum_M DTWL
MUL_4138|M.ulcerans_Agy99 DTWL
MAV_0594|M.avium_104 DTWL
MSMEG_6035|M.smegmatis_MC2_155 DTWL
TH_1739|M.thermoresistible__bu DTWL
MAB_0586|M.abscessus_ATCC_1997 DTWL
MLBr_02561|M.leprae_Br4923 ----