For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGSPPIDPRTFRNVLGQFCTGITVITTVHDDAPVGFACQSFAALSLDPPLVLFCPTKASRSWAAIEASGT FCVNILHENQQHVSARFGSREPDKFAGIDWAPSKLGSPVIEGSLAHIDCTVHSVHDGGDHFVVFGAVHSL SDVPKKKPRPLLFYHGQYTGIEPDKNTPADWRNDLEAFLTATTDDTWL
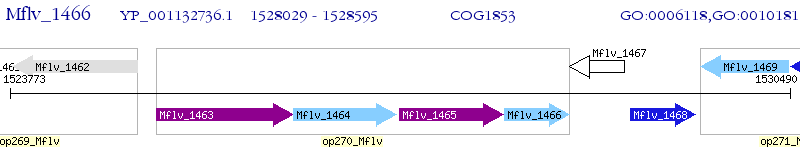
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1466 | - | - | 100% (188) | flavin reductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_4569 | - | 1e-34 | 42.95% (156) | flavin reductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0399 | - | 8e-17 | 28.48% (151) | flavin reductase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3598c | - | 2e-91 | 83.33% (186) | oxidoreductase |
| M. tuberculosis H37Rv | Rv3567c | - | 9e-91 | 82.80% (186) | oxidoreductase |
| M. leprae Br4923 | MLBr_02561 | - | 3e-15 | 24.84% (153) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0586 | - | 5e-88 | 78.24% (193) | oxidoreductase |
| M. marinum M | MMAR_5062 | - | 8e-94 | 83.51% (188) | oxidase |
| M. avium 104 | MAV_0594 | - | 9e-93 | 83.15% (184) | putative flavin reductase |
| M. smegmatis MC2 155 | MSMEG_6035 | - | 5e-98 | 87.50% (184) | nitrilotriacetate monooxygenase component B |
| M. thermoresistible (build 8) | TH_1739 | - | 8e-95 | 83.51% (188) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4138 | - | 4e-93 | 82.98% (188) | oxidase |
| M. vanbaalenii PYR-1 | Mvan_5305 | - | 1e-105 | 92.55% (188) | flavin reductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3598c|M.bovis_AF2122/97 -MSA-QIDPRTFRSVLGQFCTGITVITTVHDDVPVGFACQSFAALSLEPP
Rv3567c|M.tuberculosis_H37Rv -MSA-QIDPRTFRSVLGQFCTGITVITTVHDDVPVGFACQSFAALSLEPP
MMAR_5062|M.marinum_M -MAADPIDPRTFRHVLGQFCTGITIITTVHDDVPSGFACQSFAALSLDPP
MUL_4138|M.ulcerans_Agy99 -MAADPIDPRTFRHVLGQFCTGITIITTVHDDVPSGFACQSFAALSLDPP
MAV_0594|M.avium_104 -MAA-PIDPRTFRQVLGQFCTGITIITTVHDEVPVGFACQSFAALSLDPP
Mflv_1466|M.gilvum_PYR-GCK -MGSPPIDPRTFRNVLGQFCTGITVITTVHDDAPVGFACQSFAALSLDPP
Mvan_5305|M.vanbaalenii_PYR-1 -MSKPPIDPRTFRNVLGQFCTGITVITTVHDGSPVGFACQSFAALSLDPP
MSMEG_6035|M.smegmatis_MC2_155 MTATQPIDPRTFRNVLGQFCTGVTVITTVHENVPVGFACQSFAALSLDPP
TH_1739|M.thermoresistible__bu -VTAEAIDQRTFRRVLGQFCTGVTIITTVHEGNPVGFACQSFAALSLDPP
MAB_0586|M.abscessus_ATCC_1997 -MSTPEIDPRTFRNVLGQFCTGITIITTTHEDVPVGFACQSFAALSLEPP
MLBr_02561|M.leprae_Br4923 MRAMNSLTPSLLRDAFGHFPSGVVAIAAEVEGVREGLAASTFVPVSLEPP
: :* .:*:* :*:. *:: : *:*..:*..:**:**
Mb3598c|M.bovis_AF2122/97 LVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFGSKEPDKFAGID
Rv3567c|M.tuberculosis_H37Rv LVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFGSKEPDKFAGID
MMAR_5062|M.marinum_M LVLFCPTKVSRSWKAIEASGRFCVNVLTEKQKSVSARFGSKEPDKFAGID
MUL_4138|M.ulcerans_Agy99 LVLFCPTKVSRSWKAIEASGRFCVNVLTEKQKSVSARFGSKESDKFAGID
MAV_0594|M.avium_104 LVLFCPTKVSRSWKAIEATGRFCVNVLTEKQKHVSARFGSKEPDKFAGID
Mflv_1466|M.gilvum_PYR-GCK LVLFCPTKASRSWAAIEASGTFCVNILHENQQHVSARFGSREPDKFAGID
Mvan_5305|M.vanbaalenii_PYR-1 LVLFCPTKASRSWAAIEASGTFCVNILHENQKDVSARFGSREPDKFAGID
MSMEG_6035|M.smegmatis_MC2_155 LVLFCPTKQSRAWKAIEATGRFCVNMLHENQQHVSAQFGSKAADKFAGID
TH_1739|M.thermoresistible__bu LVLFCPTKVSRSWKAIEASGRFCVNILHEKQQHVSARFGSREPDKFAGID
MAB_0586|M.abscessus_ATCC_1997 LVLFCPTKQSRSWAAIAASGKFCVNVLAEDQREVSARFGSREPDKFAGIN
MLBr_02561|M.leprae_Br4923 LVSFCVQNVSTTWLKLKAVPMLGISVLGEVHDEVACTLATKHGDRFAGLE
** ** : * :* : * : :.:* * : *:. :.:: *:***::
Mb3598c|M.bovis_AF2122/97 WRPSELGSPIIEGSLAYIDCTVASVHDGGDHFVVFGAVESLSE---VPAV
Rv3567c|M.tuberculosis_H37Rv WRPSELGSPIIEGSLAYIDCTVASVHDGGDHFVVFGAVESLSE---VPAV
MMAR_5062|M.marinum_M WRPSELGSPIIDGSLAYIDCTVASVHDGGDHFVVFGAVHSLSE---VPKI
MUL_4138|M.ulcerans_Agy99 WRPSELGSPIIDGSLAYIDCTVASVHDGGDHFVVFGAVHSLSE---VPKI
MAV_0594|M.avium_104 WHPSELGSPIIDGALAHIDCTVASVHEGGDHFVVFGAVQSLSE---APKI
Mflv_1466|M.gilvum_PYR-GCK WAPSKLGSPVIEGSLAHIDCTVHSVHDGGDHFVVFGAVHSLSD---VPKK
Mvan_5305|M.vanbaalenii_PYR-1 WTPSKTGSPIIDGTLAHIDCSVHSVHDGGDHFVVFGAVHSLSD---VPKT
MSMEG_6035|M.smegmatis_MC2_155 WHPSKLGSPVIDGTLAHIDCTVHSVHDGGDHFVVFGAVHSLSE---VPKT
TH_1739|M.thermoresistible__bu WRPSDLGSPIIDGSLAHIDCTVHDVHDGGDHFVVFGKVHGLSE---VPER
MAB_0586|M.abscessus_ATCC_1997 WRPSGLGSPIINGSLAHIDCEVANVHDGGDHWVVFGSVKELSERAVIPSA
MLBr_02561|M.leprae_Br4923 TVSNDTGAVFIKGTSLWLESAIKQLIPAGDHTIVVLRVSQVTVD---PDV
.. *: .*.*: ::. : .: .*** :*. * :: *
Mb3598c|M.bovis_AF2122/97 K--PRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLTTTTQDTWL
Rv3567c|M.tuberculosis_H37Rv K--PRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLITTTQDTWL
MMAR_5062|M.marinum_M K--PRPLLFYRGDYTGIEPDKTTPAQWRDDLEAFLTATTQDTWL
MUL_4138|M.ulcerans_Agy99 K--PRPLLFYRGDYTGIEPDKTTPAQWRDDLEAFLTATTQDTWL
MAV_0594|M.avium_104 K--PRPLLFYRGEYTGIEPDKTTPAQWRDDLEAFLTTTTQDTWL
Mflv_1466|M.gilvum_PYR-GCK K--PRPLLFYHGQYTGIEPDKNTPADWRNDLEAFLTATTDDTWL
Mvan_5305|M.vanbaalenii_PYR-1 K--PRPLLFYQGQYTGIEPDKNTPADWRNDLEAFLTATTDDTWL
MSMEG_6035|M.smegmatis_MC2_155 K--PRPLLFYRGEYTGIEPDKNTPAQWRDDLEAFLTATTDDTWL
TH_1739|M.thermoresistible__bu K--PRPLLFYRGEYTGIEPEKNTPAQWRDDLEAFLTATTADTWL
MAB_0586|M.abscessus_ATCC_1997 ESAARPLLFYRGQYTGIEPDKNTPADWRNDLEAFLTATTEDTWL
MLBr_02561|M.leprae_Br4923 A----PIVFHRSMFRRLGF-------------------------
*::*::. : :