For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVEAFVLVQTEVGRAEVIAKQLAGLTGVLSAEYVTGPYDVVVRVGAPSLDDLKSGVVPAVQQVAGITRTL TCPIADGAAP
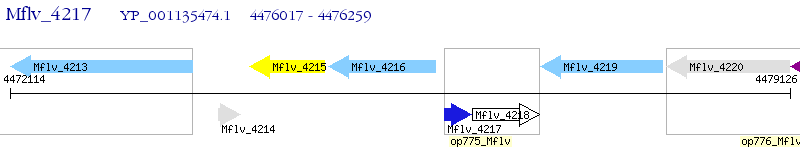
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4217 | - | - | 100% (80) | AsnC family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0892c | - | 5e-05 | 35.14% (74) | AsnC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2397 | - | 2e-29 | 77.92% (77) | transcriptional regulatory protein, AsnC family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2150 | - | 5e-35 | 90.00% (80) | AsnC family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4217|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_2150|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_2397|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0892c|M.abscessus_ATCC_199 MAGSGEFDNLDQALITAMYAHPRAGALELSRNLHVARATVQARLEKLEDS
Mflv_4217|M.gilvum_PYR-GCK -----------------MVEAFVLVQTEVGRAEVIAKQLAGLTGVLSAEY
Mvan_2150|M.vanbaalenii_PYR-1 -----------------MVEAFVLIQTEVGRAEVIAKQLAGLTGVVSAEY
MSMEG_2397|M.smegmatis_MC2_155 -----------------MVEAYVLIQTEVGRAEVVAKQLASLAGVVSAEY
MAB_0892c|M.abscessus_ATCC_199 GVIEGYEPRINLAAAGFGVQAFVTLEIAQGALSRVSDELAAIPGVLEAYA
*:*:* :: * . ::.:**.:.**:.*
Mflv_4217|M.gilvum_PYR-GCK VTGPYDVVVRVGAPSLDDLK--------SGVVPAVQQVAGITRTLT---C
Mvan_2150|M.vanbaalenii_PYR-1 VTGPYDVVVRVTANSLDELK--------STVVPSVQQVAGITRTLT---C
MSMEG_2397|M.smegmatis_MC2_155 VTGPYDVVLRVSADTLEALR--------ADVVPSVQQVAGITRTLT---C
MAB_0892c|M.abscessus_ATCC_199 TTGDGDVLCRVAAESHDRLQQTLLELNQSGVVQRSTSVIALSVVVPPRVL
.** **: ** * : : *: : ** .* .:: .:.
Mflv_4217|M.gilvum_PYR-GCK PIADG---AAP------------
Mvan_2150|M.vanbaalenii_PYR-1 PIADG---ARP------------
MSMEG_2397|M.smegmatis_MC2_155 PIASG---T--------------
MAB_0892c|M.abscessus_ATCC_199 PLLQSREVDRPATRAPAYRKTQQ
*: ..