For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGAVCPGSFDPVTLGHIDVFERAAAQFDEIVVAVMVNPNKSGMFTLDERIALIEESTTHLPNLRVESGQG LIVDFVRERGLTAIVKGLRTGTDFEYELQMAQMNKHVAGIDTFFVATAPSYSFVSSSLAKEVAMLGGDVT ALLPAAVNTRLTAKLAERG*
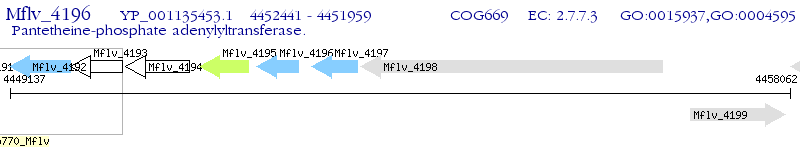
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4196 | coaD | - | 100% (160) | phosphopantetheine adenylyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2989c | coaD | 2e-71 | 80.77% (156) | phosphopantetheine adenylyltransferase |
| M. tuberculosis H37Rv | Rv2965c | coaD | 2e-71 | 80.77% (156) | phosphopantetheine adenylyltransferase |
| M. leprae Br4923 | MLBr_01663 | coaD | 2e-68 | 79.08% (153) | phosphopantetheine adenylyltransferase |
| M. abscessus ATCC 19977 | MAB_3259c | coaD | 3e-73 | 83.33% (156) | phosphopantetheine adenylyltransferase |
| M. marinum M | MMAR_1751 | coaD | 1e-71 | 83.55% (152) | phosphopantetheine adenylyltransferase KdtB |
| M. avium 104 | MAV_3810 | coaD | 3e-73 | 82.39% (159) | phosphopantetheine adenylyltransferase |
| M. smegmatis MC2 155 | MSMEG_2414 | coaD | 9e-76 | 87.82% (156) | phosphopantetheine adenylyltransferase |
| M. thermoresistible (build 8) | TH_1227 | kdtB | 1e-76 | 84.38% (160) | PROBABLE PHOSPHOPANTETHEINE ADENYLYLTRANSFERASE KDTB |
| M. ulcerans Agy99 | MUL_1994 | coaD | 9e-72 | 83.55% (152) | phosphopantetheine adenylyltransferase |
| M. vanbaalenii PYR-1 | Mvan_2167 | coaD | 1e-76 | 87.42% (159) | phosphopantetheine adenylyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4196|M.gilvum_PYR-GCK MSGAVCPGSFDPVTLGHIDVFERAAAQFDEIVVAVMVNPNKSGMFTLDER
Mvan_2167|M.vanbaalenii_PYR-1 MSGAVCPGSFDPVTLGHIDIFERAAAQFDEVVVAVMVNPNKTGMFTHEER
MMAR_1751|M.marinum_M MTGAVCPGSFDPVTLGHVDVFERAAAQFDEVVVAILVNPAKKGMFDLDER
MUL_1994|M.ulcerans_Agy99 MTGAVCPGSFDPVTLGHVDVFERAAAQFDEVVVAILVNPAKKGMFDLDER
Mb2989c|M.bovis_AF2122/97 MTGAVCPGSFDPVTLGHVDIFERAAAQFDEVVVAILVNPAKTGMFDLDER
Rv2965c|M.tuberculosis_H37Rv MTGAVCPGSFDPVTLGHVDIFERAAAQFDEVVVAILVNPAKTGMFDLDER
MLBr_01663|M.leprae_Br4923 ---MVCPGSFDPVTLGHIDVFERAAAQFDEVVVAILINPVKKGMFDLDER
MAV_3810|M.avium_104 MTGAVCPGSFDPVTLGHVDVFERASAQFDEVVVAILTNPAKKGMFDLDER
TH_1227|M.thermoresistible__bu MSGAVCPGSFDPVTLGHVDVFERAAAQFDEVIVAVLVNPNKQGMFSLEER
MSMEG_2414|M.smegmatis_MC2_155 MSGAVCPGSFDPVTLGHIDVFERASAQFDEVVVAVLVNPNKKGMFDLDER
MAB_3259c|M.abscessus_ATCC_199 MTGAVCPGSFDPVTLGHLDVFERAAAQFDEVIVAVLINPNKAGMFTVDER
*************:*:****:*****::**:: ** * *** :**
Mflv_4196|M.gilvum_PYR-GCK IALIEESTTHLPNLRVESGQGLIVDFVRERGLTAIVKGLRTGTDFEYELQ
Mvan_2167|M.vanbaalenii_PYR-1 IALIEESTTHLPNLRVESGQGLIVDFVKARGLTAIVKGLRTGTDFEYELQ
MMAR_1751|M.marinum_M IAMIEESTAHLPNLRVEAGQGLVVDFVKAQGMNAIVKGLRTGTDFEYELQ
MUL_1994|M.ulcerans_Agy99 IAMIEESTAHLPNLRVEAGQGLVVDFVKAQGMNAIVKGLRTGTDFEYELQ
Mb2989c|M.bovis_AF2122/97 IAMVKESTTHLPNLRVQVGHGLVVDFVRSCGMTAIVKGLRTGTDFEYELQ
Rv2965c|M.tuberculosis_H37Rv IAMVKESTTHLPNLRVQVGHGLVVDFVRSCGMTAIVKGLRTGTDFEYELQ
MLBr_01663|M.leprae_Br4923 IAMINESTMHLPNLRVEAGEGLVVALVRSRGMTAIVKGLRTGVDFEYELQ
MAV_3810|M.avium_104 IAMIEESTTHLSNLRVEAGQGLVVDFVRSRGMTAIVKGLRTGTDFEYELQ
TH_1227|M.thermoresistible__bu IEMIEESTTHLPNLRVESGSGLVVDFVKSRGMTAIVKGLRTGTDFEYELQ
MSMEG_2414|M.smegmatis_MC2_155 IAMIEESTTHLPNLRVESGQGLVVDFVKSRGLTAIVKGLRTGTDFEYELQ
MAB_3259c|M.abscessus_ATCC_199 IEMIRESTADLPNLRVESGQGLLVDFVRERGLNAIVKGLRTGTDFEYELQ
* ::.*** .*.****: * **:* :*: *:.*********.*******
Mflv_4196|M.gilvum_PYR-GCK MAQMNKHVAGIDTFFVATAPSYSFVSSSLAKEVAMLGGDVTALLPAAVNT
Mvan_2167|M.vanbaalenii_PYR-1 MAQMNNHIAGVDTFFIATTPRYSFVSSSLAKEVATLGGDVSELLPAPVNA
MMAR_1751|M.marinum_M MAQMNKHIAGVDTFFVATAPRYSFVSSSLAKEVAMLGGDVSELLPEPVNR
MUL_1994|M.ulcerans_Agy99 MAQMNKHIAGVDTFFVATAPRYSFVSSSLAKEVAMLGGDVSELLPEPVNR
Mb2989c|M.bovis_AF2122/97 MAQMNKHIAGVDTFFVATAPRYSFVSSSLAKEVAMLGGDVSELLPEPVNR
Rv2965c|M.tuberculosis_H37Rv MAQMNKHIAGVDTFFVATAPRYSFVSSSLAKEVAMLGGDVSELLPEPVNR
MLBr_01663|M.leprae_Br4923 MAQMNKHIAGVDTFFVATAPRYSFVSSSLVKEVAMLGGDVSELLPESVNR
MAV_3810|M.avium_104 MAQMNKHIAGVDTFFVATAPRYSFVSSSLAKEVAMLGGDVSELLPEPVNR
TH_1227|M.thermoresistible__bu MAQMNKHIAGVDTFFVATTPRYSFVSSSLAKEVAMLGGDVSELLPGPVNA
MSMEG_2414|M.smegmatis_MC2_155 MAQMNKHVAGVDTFFVATTPQYSFVSSSLAKEVASLGGDVSALLPSPVNR
MAB_3259c|M.abscessus_ATCC_199 MAQMNKHIAGVDTFFVATAPAYSFVSSSLAKEVATYGGDVSALLPASVHQ
*****:*:**:****:**:* ********.**** ****: *** .*:
Mflv_4196|M.gilvum_PYR-GCK RLTAKLAERG-
Mvan_2167|M.vanbaalenii_PYR-1 RLKAKLAARD-
MMAR_1751|M.marinum_M RLRERTK----
MUL_1994|M.ulcerans_Agy99 RLRERTK----
Mb2989c|M.bovis_AF2122/97 RLRDRLNTERT
Rv2965c|M.tuberculosis_H37Rv RLRDRLNTERT
MLBr_01663|M.leprae_Br4923 RFREKMSGTS-
MAV_3810|M.avium_104 RLREKLSGRS-
TH_1227|M.thermoresistible__bu RLRKKLAERG-
MSMEG_2414|M.smegmatis_MC2_155 RLQEKLNG---
MAB_3259c|M.abscessus_ATCC_199 RLLGKLRGQAQ
*: :