For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AGYYICPVEDQQLVDRASSALAEVDTPGWAQRFDLVSDPHRLEILLCLHRAPGICVSDLAAALGRSENTV SQALRVLRQQGWVTSTRAGRSVTYRLEDEVVHDLLHWIGARHG*
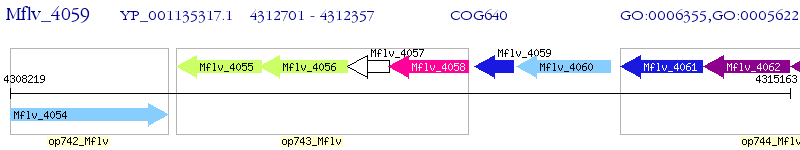
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4059 | - | - | 100% (114) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_2832 | - | 2e-09 | 44.00% (75) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_1658 | - | 2e-08 | 38.64% (88) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3770 | - | 9e-10 | 43.18% (88) | transcriptional regulatory protein ArsR-family |
| M. tuberculosis H37Rv | Rv3744 | - | 9e-10 | 43.18% (88) | transcriptional regulatory protein ArsR-family |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1679c | - | 2e-06 | 42.62% (61) | transcription regulator ArsR |
| M. marinum M | MMAR_3668 | - | 3e-07 | 45.90% (61) | ArsR family transcriptional regulator |
| M. avium 104 | MAV_1492 | - | 6e-08 | 41.10% (73) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_2606 | - | 6e-42 | 78.00% (100) | ArsR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1389 | - | 4e-44 | 77.88% (104) | ArsR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_3611 | - | 2e-07 | 45.90% (61) | ArsR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_2283 | - | 3e-56 | 91.15% (113) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3770|M.bovis_AF2122/97 ----------------------MGHGVEGRNRPSAPLDSQAAAQVASTLQ
Rv3744|M.tuberculosis_H37Rv ----------------------MGHGVEGRNRPSAPLDSQAAAQVASTLQ
Mflv_4059|M.gilvum_PYR-GCK ---------------MAGYYICPVEDQQLVDRASSALAEVDTPGWAQRFD
Mvan_2283|M.vanbaalenii_PYR-1 MSARRHAADAGVLRAPAADYIARVDDQQLVERATSALAEVDTPRWAQRFD
MSMEG_2606|M.smegmatis_MC2_155 -----------------------------MERATSALVDVDSATWARRFD
TH_1389|M.thermoresistible__bu --------------------VDVSGDQLALEHASAALADVDTTEWSRRFD
MMAR_3668|M.marinum_M -MATSPSPSTAAHDDFVSRHEHDVGDFAEHPAFPAPPPREILEGAGELLR
MUL_3611|M.ulcerans_Agy99 -MATSPSPSTAAHDDFVSRHEHDVGDFAEHPAFPAPPPREILEGAGELLR
MAB_1679c|M.abscessus_ATCC_199 -MVN--TVLSSAHDE--PPHAPLT------PPGPLPP-REILEQSGELLR
MAV_1492|M.avium_104 -------------MPKTLPVIDMSEPVCCAPVAAGPADDAAALDVAMRLK
. . . :
Mb3770|M.bovis_AF2122/97 ALATPSRLMILTQLRNGP-LPVTD-LAEAIGMEQSAVSHQLRVLRNLGLV
Rv3744|M.tuberculosis_H37Rv ALATPSRLMILTQLRNGP-LPVTD-LAEAIGMEQSAVSHQLRVLRNLGLV
Mflv_4059|M.gilvum_PYR-GCK LVSDPHRLEILLCLHRAPGICVSD-LAAALGRSENTVSQALRVLRQQGWV
Mvan_2283|M.vanbaalenii_PYR-1 LVSDPHRLEILLCLHRAPGICVSD-LAAALGRSENTVSQALRVLRQQGWV
MSMEG_2606|M.smegmatis_MC2_155 LLSDPNRLEILLVLHRAPGIFVGD-LAEVLGRSENAVSQALRVLRQQGWV
TH_1389|M.thermoresistible__bu LLSDPHRLEILLVLHRAPGIRVGD-LAAALGRSENAVSQALRVLRQQGWV
MMAR_3668|M.marinum_M ALAAPVRIAIVLQLREAQ-RCVHE-LVDALGVPQPLVSQHLKILKAAGVV
MUL_3611|M.ulcerans_Agy99 ALAAPVRIAIVLQLREAQ-RCVHE-LVDALGVPQPLVSQHLKILKAAGVV
MAB_1679c|M.abscessus_ATCC_199 ALAAPVRIAIVLQLRESQ-RCVHE-LVDAVGVAQPLISQHLRVLKAAGVV
MAV_1492|M.avium_104 ALADPVRVKLMSLLFTADGTCTTGSLAAAVGLSESTVSHHLGQLRTAGFV
:: * *: :: * . . *. .:* : :*: * *: * *
Mb3770|M.bovis_AF2122/97 VGDRAGRSIVYSLYDTHVAQLLDEAIYHSEHLHLGLSDRHPSAG
Rv3744|M.tuberculosis_H37Rv VGDRAGRSIVYSLYDTHVAQLLDEAIYHSEHLHLGLSDRHPSAG
Mflv_4059|M.gilvum_PYR-GCK TSTRAGRSVTYRLEDEVVHDLLH-----------WIGARHG---
Mvan_2283|M.vanbaalenii_PYR-1 TNTRAGRSVTYRLEDETVHDLLH-----------WIGARHG---
MSMEG_2606|M.smegmatis_MC2_155 SSTRVGRAVSYRLEDDVVHELLH-----------WIGARHG---
TH_1389|M.thermoresistible__bu RSTRIGRTVSYRLDDDIVHDLLH-----------WIGARHG---
MMAR_3668|M.marinum_M TGERSGREVLYRLVDQHLAHIVVD----------AVAHAGEDSA
MUL_3611|M.ulcerans_Agy99 TGERSGREVLYRLVDQHLAHIVVD----------AVAHAGEDSA
MAB_1679c|M.abscessus_ATCC_199 SGERQGREVMYRLVDDHLSHIVVD----------AVAHTEEQR-
MAV_1492|M.avium_104 ASDRQGMSVHHTARREALTALCR-----------VLDPSCCT--
. * * : : : : :