For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NRQTSRQSKWPSPKLLTGCPRVDGRTRLARNRCGADSVTLPSAIATHFDICQYRPMSKSLGVLHMGGSLG RAALDEAACVPLAQMFKALGDPARLRLLSLIASNPGGEACVCDIAASFDLSQPTISHHLKVLRTAGILDS ERRGSWVYYRAIPEALQRLSTILHVDGGTPFPSDACSEETAR*
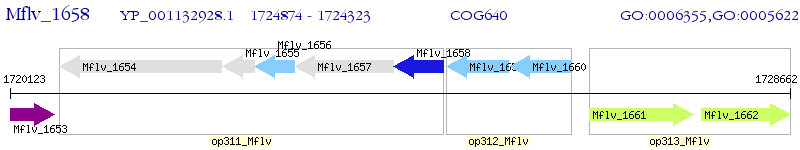
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1658 | - | - | 100% (183) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_3471 | - | 9e-18 | 39.17% (120) | regulatory protein, ArsR |
| M. gilvum PYR-GCK | Mflv_1652 | - | 3e-16 | 38.79% (116) | regulatory protein, ArsR |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2675 | - | 9e-29 | 61.86% (97) | ArsR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv2642 | - | 9e-29 | 61.86% (97) | ArsR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4862 | - | 3e-30 | 60.40% (101) | ArsR family transcriptional regulator |
| M. marinum M | MMAR_2056 | - | 1e-29 | 59.60% (99) | ArsR family transcriptional regulator |
| M. avium 104 | MAV_1492 | - | 4e-13 | 42.70% (89) | putative transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_1173 | - | 9e-28 | 56.00% (100) | transcriptional regulator, ArsR family protein |
| M. thermoresistible (build 8) | TH_1047 | - | 3e-31 | 54.26% (129) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN (PROBABLY |
| M. ulcerans Agy99 | MUL_3287 | - | 7e-30 | 59.60% (99) | ArsR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_3223 | - | 5e-30 | 58.25% (103) | regulatory protein, ArsR |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1173|M.smegmatis_MC2_155 --------------------------------------------------
TH_1047|M.thermoresistible__bu --------------------------------MFTTRSLDDGFDDYQYRY
Mvan_3223|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2675|M.bovis_AF2122/97 --------------------------------------------------
Rv2642|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_2056|M.marinum_M --------------------------------------------------
MUL_3287|M.ulcerans_Agy99 --------------------------------------------------
MAB_4862|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_1658|M.gilvum_PYR-GCK MNRQTSRQSKWPSPKLLTGCPRVDGRTRLARNRCGADSVTLPSAIATHFD
MAV_1492|M.avium_104 --------------------------------------------------
MSMEG_1173|M.smegmatis_MC2_155 MSN-QIAATDLAACCPPGALLREPLSAAEAADMSVKLKALADPVRLQLFS
TH_1047|M.thermoresistible__bu MSKSLWESIDGAATCPPAALLREPLSAAEAADVATTLKALADPVRLQLFS
Mvan_3223|M.vanbaalenii_PYR-1 MSKSAAPRAD-DQCCPPGALLRKPLSAAAAADMAVRFKALADPVRLQLFS
Mb2675|M.bovis_AF2122/97 MSN--LHPLPEVASCVVAPLVREPLNPPAAAEMAARFKALADPVRLQLLS
Rv2642|M.tuberculosis_H37Rv MSN--LHPLPEVASCVVAPLVREPLNPPAAAEMAARFKALADPVRLQLLS
MMAR_2056|M.marinum_M MSN--RVEM-DAPCCVAAPLLREPLEASSAVEMAARFKALADPVRLQLLS
MUL_3287|M.ulcerans_Agy99 MSN--RVEM-DAPCCVTAPLLREPLEASSAVEMAARFKALADPVRLQLLS
MAB_4862|M.abscessus_ATCC_1997 MSN---QLVIEPVNIRCSPLVREPISAGQAVDLAKVFKALGDPVRLRLFS
Mflv_1658|M.gilvum_PYR-GCK ICQYRPMSKSLGVLHMGGSLGRAALDEAACVPLAQMFKALGDPARLRLLS
MAV_1492|M.avium_104 MPK-TLPVIDMSEPVCCAPVAAGPADDAAALDVAMRLKALADPVRVKLMS
: : ..: . . . :: :***.**.*::*:*
MSMEG_1173|M.smegmatis_MC2_155 AVASREGGEACVCDIAAGVEVSQPTVSHHLKVLRDAGLLTSQRRASWVYY
TH_1047|M.thermoresistible__bu AIASHAGGEACVCDISTGLEVSQPTVSHHLKVLREAGLLTSERRGSWVYY
Mvan_3223|M.vanbaalenii_PYR-1 AIASHDGGEACVCDLSAGIEVSQPTVSHHLKVLRDAGLLTSQRRASWVYY
Mb2675|M.bovis_AF2122/97 SVASRAGGEACVCDISAGVEVSQPTISHHLKVLRDAGLLTSRRRASWVYY
Rv2642|M.tuberculosis_H37Rv SVASRAGGEACVCDISAGVEVSQPTISHHLKVLRDAGLLTSRRRASWVYY
MMAR_2056|M.marinum_M SVASHAGGEACVCDISAGVEVGQPTISHHLKVLRDAGLLTSQRRASWVYY
MUL_3287|M.ulcerans_Agy99 SVASHAGGEACVCDISAGVEVGQPTISHHLKVLRDAGLLTSQRRASWVYY
MAB_4862|M.abscessus_ATCC_1997 LIASHAGGEACVCDISPGIEVSQPTISHHLKVLRNAGLLASERRASWVYY
Mflv_1658|M.gilvum_PYR-GCK LIASNPGGEACVCDIAASFDLSQPTISHHLKVLRTAGILDSERRGSWVYY
MAV_1492|M.avium_104 LLFTADG-TCTTGSLAAAVGLSESTVSHHLGQLRTAGFVASDRQGMSVHH
: : * . . .::... :.:.*:**** ** **:: * *:. *::
MSMEG_1173|M.smegmatis_MC2_155 AVVPDALASLAVLLGADLRAEVTA---------
TH_1047|M.thermoresistible__bu AVVPQALASLSALLGATGTAPVGAPR-------
Mvan_3223|M.vanbaalenii_PYR-1 AVVPEALDSLAVILGAD-AASVPA---------
Mb2675|M.bovis_AF2122/97 AVVPEALTVLSNLLSVHADAAPALGAPA-----
Rv2642|M.tuberculosis_H37Rv AVVPEALTVLSNLLSVHADAAPALGAPA-----
MMAR_2056|M.marinum_M AVVPEALNALSRLLSVHAGSATELETSA-----
MUL_3287|M.ulcerans_Agy99 AVVPEALNALSRLLSVHAGSATELETSA-----
MAB_4862|M.abscessus_ATCC_1997 QVVPEVLDALAGIVRIPDTTISATPTEGPS---
Mflv_1658|M.gilvum_PYR-GCK RAIPEALQRLSTILHVDGGTPFPSDACSEETAR
MAV_1492|M.avium_104 TARREALTALCRVLDPSCCT-------------
. :.* *. :: :