For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSVSASPGRRRGDLDGLRGVAIALVAVFHIWFGRVSGGVDVFLVLSGYFFGAMVLGRALTPGASLSPLPM LRRLARRLLPALIVVLGGAAVLTFTLQPQTRWESFADQSLASLGYYQNWYLSAAASDYARAGEAISPLQH LWSMSVQGQFYLAMLAVTLGLAAVLRRFVGSRLKAVFVGVFVVLAAASFWYAIVAHGEHQAGAYYDSFAR AWELLAGVLAALLLTRVSMPMWARTAAATVGVVAILSCGALIDGAQLFPGPWALVPVGATVLVIAAGAGQ PRREPAPLRLLAGRPAVALGAMAYSLYLWHWPVLIFWLVHTGAPRVGILDGLGVLAVSLVLAYLTTRLIE NPLRHPDSAGDADRAARVWVWVRRPRFALGSAVVLLAVSLAVTSFAWREHVVMQRAKGHELAQLPERDYP GALALTNRIRVPQLPMRPTVLEARDDIPATTLDGCISDFVTVELINCIYGDPDAVRTIALAGGSHAEHWI TALDTLGTRHGFRVVTYLKMGCPLTTEMVPTIAISSEEYPQCHEWNQRAMDKLLTDRPDFVFTTTTRPNA DEPGDLVPEAYLGIWDTLSANGISILGMRDTPWLIRDGRLADPADCLSRGGDSVSCGVKRSWALSETNPT LDLASYYPLLHPLDMSDAVCDETDCRVTEGNILVYHDSHHVSATYMRTMTAELGRQIAAATGWW
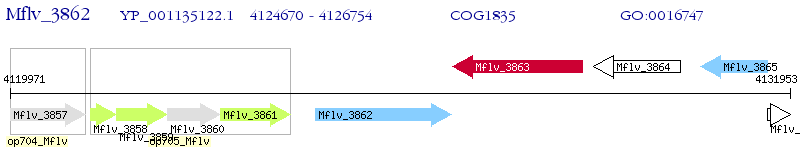
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3862 | - | - | 100% (694) | acyltransferase 3 |
| M. gilvum PYR-GCK | Mflv_3630 | - | 0.0 | 62.55% (697) | acyltransferase 3 |
| M. gilvum PYR-GCK | Mflv_2821 | - | 0.0 | 58.61% (703) | acyltransferase 3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1592c | - | 0.0 | 62.91% (701) | hypothetical protein Mb1592c |
| M. tuberculosis H37Rv | Rv1565c | - | 0.0 | 62.91% (701) | hypothetical protein Rv1565c |
| M. leprae Br4923 | MLBr_01213 | - | 0.0 | 59.94% (694) | hypothetical protein MLBr_01213 |
| M. abscessus ATCC 19977 | MAB_2689 | - | 0.0 | 59.59% (688) | acyltransferase |
| M. marinum M | MMAR_2380 | - | 0.0 | 60.55% (697) | hypothetical protein MMAR_2380 |
| M. avium 104 | MAV_3209 | - | 0.0 | 62.54% (702) | acyltransferase domain-containing protein |
| M. smegmatis MC2 155 | MSMEG_3187 | - | 0.0 | 63.06% (693) | acyltransferase domain-containing protein |
| M. thermoresistible (build 8) | TH_1799 | - | 0.0 | 60.83% (697) | CONSERVED HYPOTHETICAL MEMBRANE PROTEIN |
| M. ulcerans Agy99 | MUL_1556 | - | 0.0 | 60.26% (697) | hypothetical protein MUL_1556 |
| M. vanbaalenii PYR-1 | Mvan_2787 | - | 0.0 | 62.45% (695) | acyltransferase 3 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_3187|M.smegmatis_MC2_155 -------MTLSPTRPAP--ISRDTRAANP----SMGTRKSG---FYRHDL
Mvan_2787|M.vanbaalenii_PYR-1 ------MMTLAPARPASPTVPGPTRTASVPPSAGMGTRTKG---FYRHDL
TH_1799|M.thermoresistible__bu ---------------------------------VTGPGTSG---FYRYDL
Mb1592c|M.bovis_AF2122/97 ---------MLTLSPPRPPALTPEPALPP---VTMGTRTTG---FYRHDL
Rv1565c|M.tuberculosis_H37Rv ---------MLTLSPPRPPALTPEPALPP---VTMGTRTTG---FYRHDL
MMAR_2380|M.marinum_M ---------MQTLTPPTP----PTAAAAP---VAMGTRKTG---FYRHDL
MUL_1556|M.ulcerans_Agy99 ---------MQTLTPPTS----PTAAAAP---VAMGTRKTG---FYRHDL
MLBr_01213|M.leprae_Br4923 MSSVNWRSIMQTLSPASL----LVATAKP---VPLEGRTAVFTIFYRHDL
MAV_3209|M.avium_104 ---------MQTPSPSPA----PVAAADP---GTSGARTPGSRGFYRYDL
MAB_2689|M.abscessus_ATCC_1997 ---------MFFVSATKPTKDPEVSPAAA---MDATPKPKGDKAFYRYDL
Mflv_3862|M.gilvum_PYR-GCK ----------------------MSVSASP-------GRRRG-------DL
**
MSMEG_3187|M.smegmatis_MC2_155 DGLRGIAIMMVAVFHIWFGRVSGGVDVFLALSGFFFGGKILRTALDQSTP
Mvan_2787|M.vanbaalenii_PYR-1 DGLRGIAIALVAVFHVWFGRVSGGVDVFLVLSGFFFGGRLLRGALTPGVA
TH_1799|M.thermoresistible__bu DGLRGVAILLVALFHVWFGRVSGGVDVFLALSGFFFGGRLLRAVLAPGAS
Mb1592c|M.bovis_AF2122/97 DGLRGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNPDLS
Rv1565c|M.tuberculosis_H37Rv DGLRGVAIALVAVFHVWFGRVSGGVDVFLALSGFFFGGKILRAALNPDLS
MMAR_2380|M.marinum_M DGLRGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNPSLK
MUL_1556|M.ulcerans_Agy99 DGLRGIAIALVAIFHVWFGRVSGGVDVFLALSGFFFGGKILRTALNPSLK
MLBr_01213|M.leprae_Br4923 DGLRGIAIALVAIFHVWFGRVSGGVDVLLAMSGFFFGGKILRAALNPVPS
MAV_3209|M.avium_104 DGLRGIAIALVAVFHIWFGRVSGGVDVFLALSGFFFGGKVIRAALNPAVA
MAB_2689|M.abscessus_ATCC_1997 DGLRGIAIFLVAVFHVWFGRVSGGVDVFLTLSGFFYGSKLLRTATTQGAS
Mflv_3862|M.gilvum_PYR-GCK DGLRGVAIALVAVFHIWFGRVSGGVDVFLVLSGYFFGAMVLGRALTPGAS
*****:** :**:**:***********:*.:**:*:*. :: .
MSMEG_3187|M.smegmatis_MC2_155 LRPLSEVVRLVRRLLPALVVVLAAAAVLTILIQPETRWEAFADQSLASLG
Mvan_2787|M.vanbaalenii_PYR-1 LRPVPEVTRLIRRLLPALVVVLAVSAVLTILIQPETRWEAFADQSLASLG
TH_1799|M.thermoresistible__bu LKPLREVTRLVRRLLPALVVVLAASAVLTILIQPTTRWETFADQSLASLG
Mb1592c|M.bovis_AF2122/97 LSPIAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLASLG
Rv1565c|M.tuberculosis_H37Rv LSPIAEVIRLIRRLLPALVVVLAGCALLTIAIQPQTRWEAFANQSLASLG
MMAR_2380|M.marinum_M LSPITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLASLG
MUL_1556|M.ulcerans_Agy99 LSPITELIRLIRRLLPAMVVVLAACALLTILVQPQTRWEAFADQSLASLG
MLBr_01213|M.leprae_Br4923 LSPVAEIIRLVRRLLPALVVVLTGCALLTVVMQPQTRWETFADQSLASLG
MAV_3209|M.avium_104 LSPVAEVIRLVRRLVPALVVVLAGCAVLTILVQPETRWETFADQSLASLG
MAB_2689|M.abscessus_ATCC_1997 LNPVPVVKRLVRRLLPALILVLAACAVLTVLVQPETRWETFAEQSLASLG
Mflv_3862|M.gilvum_PYR-GCK LSPLPMLRRLARRLLPALIVVLGGAAVLTFTLQPQTRWESFADQSLASLG
* *: : ** ***:**:::** .*:**. :** ****:**:*******
MSMEG_3187|M.smegmatis_MC2_155 YYQNWELANTAADYLRAGETVSPLQHIWSMSVQGQFYIAFLVLIFGFAYL
Mvan_2787|M.vanbaalenii_PYR-1 YYQNWELARTASDYLRAGETVSPLQHIWSMSVQGQFYIAFLALIMLSAWL
TH_1799|M.thermoresistible__bu YYQNWELARTASDYLRAGEAVSPLQHIWSMSVQGQFYIAFLVLVMGFAVL
Mb1592c|M.bovis_AF2122/97 YYQNWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGCAYL
Rv1565c|M.tuberculosis_H37Rv YYQNWELASTVSNYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLVAGCAYL
MMAR_2380|M.marinum_M YYQNWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGCTFL
MUL_1556|M.ulcerans_Agy99 YYQNWELANSAADYLRAGEAVSPLQHIWSMSVQGQFYLAFLLLIAGCTFL
MLBr_01213|M.leprae_Br4923 YYQNWELVGTESSYLKAGEAVSPLQHIWSMSVQGQFDIAFLLLVAGCAYL
MAV_3209|M.avium_104 YYQNWELANTASDYLRAGEAVSPLQHIWSMSVQGQFYVSFLVLIAGLAYL
MAB_2689|M.abscessus_ATCC_1997 YYQNWELANTAADYLAASESVSPLQHLWSMSVQGQFYVGFLALVYLLAVL
Mflv_3862|M.gilvum_PYR-GCK YYQNWYLSAAASDYARAGEAISPLQHLWSMSVQGQFYLAMLAVTLGLAAV
***** * : :.* *.*::*****:********* :.:* : : :
MSMEG_3187|M.smegmatis_MC2_155 FRRVFG----RHLRTVFIVLLAALTIASFVYAIIAHNTDQATAYYNSFAR
Mvan_2787|M.vanbaalenii_PYR-1 FRRLLG----KRARAGFVVLLGALTIASFVYAIIAHNADQATAYYNSFAR
TH_1799|M.thermoresistible__bu FRRVLR----RHLRAAFVVLLTALTVASFVYAIIATNTDQATAYYNSFAR
Mb1592c|M.bovis_AF2122/97 LRRLFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNTFAR
Rv1565c|M.tuberculosis_H37Rv LRRLFRGPRAPYLRTMFVVLLSTLTLASFIYAIVAHHAYQATAYYNTFAR
MMAR_2380|M.marinum_M FRRLLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNSFAR
MUL_1556|M.ulcerans_Agy99 FRRLLG----THLRTMFVVLLSALTIASFVYAIIAHQAYQPTAYYNSFTR
MLBr_01213|M.leprae_Br4923 FRRPLG----TQLRIMFVVLLGALMIASFIYATFAHQANQTTAYYNSFAR
MAV_3209|M.avium_104 FGRPLG----ARLRTLVLVVLAALTVASFGYAIVAHQQNQAAAYYDSFAR
MAB_2689|M.abscessus_ATCC_1997 LRKPLG----RHIRTVLIVVIAVLSAASFGYAIYAHLDFQSIAYYNTFAR
Mflv_3862|M.gilvum_PYR-GCK LRRFVG----SRLKAVFVGVFVVLAAASFWYAIVAHGEHQAGAYYDSFAR
: : . : .: :: .* *** ** * *. ***::*:*
MSMEG_3187|M.smegmatis_MC2_155 AWELLLGALAGALVGFVRWPMWLRTVVSVVSLAAILSCGWFIDGVKEFPG
Mvan_2787|M.vanbaalenii_PYR-1 AWELLLGALVGAVVPHVRWPMWLRTLLAVVALAAILSCGALIDGVREFPG
TH_1799|M.thermoresistible__bu AWELLLGALAGSLVPHIRWPKWLRALAATVALAAILSCGVLIDGVNEFPG
Mb1592c|M.bovis_AF2122/97 AWELLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKEFPG
Rv1565c|M.tuberculosis_H37Rv AWELLAGALVGAVVPHVRWPMWLRTAVATAALAAILSCGALIDGVKEFPG
MMAR_2380|M.marinum_M AWELLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKEFPG
MUL_1556|M.ulcerans_Agy99 AWELLLGALVGALVPYIRWPMWLRTIAATVGLAIVLSCGALINGVKEFPG
MLBr_01213|M.leprae_Br4923 AWELLLGALVGAAVPYIRWPAWLRTVVATVALAAILSCGALINGVKEFPG
MAV_3209|M.avium_104 GWELLLGALVGALVPRIRWPMWLRTLLATVALGAVLSCGALIDGVQEFPG
MAB_2689|M.abscessus_ATCC_1997 AWELLLGVLVGALVAGTRWPRWLRQLLSFVAVVAILSCGALINGVREFPG
Mflv_3862|M.gilvum_PYR-GCK AWELLAGVLAALLLTRVSMPMWARTAAATVGVVAILSCGALIDGAQLFPG
.**** *.*.. : * * * : ..: :**** :*:*.. ***
MSMEG_3187|M.smegmatis_MC2_155 PWALVPVGATILFIFSAANRMSDPRTAGRLPAPNRLLATAPFVSLGSMAY
Mvan_2787|M.vanbaalenii_PYR-1 PWALVPVGATLLFILSAANRYADPHTRGRIPAPNRLLASKPFVALGAMAY
TH_1799|M.thermoresistible__bu PWALVPVGATILFILSAAN------WQGPLPGPNRLLATTPFVTLGSIAY
Mb1592c|M.bovis_AF2122/97 PWALVPVGATMLMILAGANRQGHPGTRDRLPLPNRLLATAPLVALGAMAY
Rv1565c|M.tuberculosis_H37Rv PWALVPVGATMLMILAGANRQGHPGTRDRLPLPNRLLATAPLVALGAMAY
MMAR_2380|M.marinum_M PWALVPVGAAMLMIMAGANRQAHPSTRDRLPLPNRLLAAGPLVTLGAMAY
MUL_1556|M.ulcerans_Agy99 PWALVPVGAAMLMIMAGANRQAHPSTRDRLPLPNRLLAAGPLVTLGAMAY
MLBr_01213|M.leprae_Br4923 PWALVPVGAAMLLILAGANRQSRPGTSASMPLPNRLLATAPLVALGTIAY
MAV_3209|M.avium_104 PWALVPVGAAMLMILVGANLAG-----DRLPLPNRLLATRPLVELGAIAY
MAB_2689|M.abscessus_ATCC_1997 PLALVPVVATLLLILSAANLPAD----ARQPVANRFLATRPLVELGALAY
Mflv_3862|M.gilvum_PYR-GCK PWALVPVGATVLVIAAGAGQPR------REPAPLRLLAGRPAVALGAMAY
* ***** *::*.* .*. * . *:** * * **::**
MSMEG_3187|M.smegmatis_MC2_155 SLYLWHWPLLIFWLSYSGHTAANFVEGAVILLVSGVLAWLTTRYIEEPLR
Mvan_2787|M.vanbaalenii_PYR-1 SLYLWHWPLLIFYLAHTGNSRVTFVEGAVILLVSGLLAWLTTRYVEEPLR
TH_1799|M.thermoresistible__bu SLYLWHWPLLIFWLAYTGNSGVNFVEGAVILLVSGVLAWLTTKLVEDPLR
Mb1592c|M.bovis_AF2122/97 SWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVEDPLR
Rv1565c|M.tuberculosis_H37Rv SWYLWHWPLLIFWLSYTGHRHANFVEGAAVLLVSGLLAYLTTRLVEDPLR
MMAR_2380|M.marinum_M SLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVEDPLR
MUL_1556|M.ulcerans_Agy99 SLYLWHWPLLIFWLSYTGHRHANFIEGAAILLVSGVLAFLTTRHVEDPLR
MLBr_01213|M.leprae_Br4923 TLYLWHWPLLIFWLSYTGHHHANFVEGAALLLVSGLLAYLTTRLIENPLR
MAV_3209|M.avium_104 SLYLWHWPLLIFWLSYSGHKHAGFVEGTVLLLVSGVLAYLTNRLVEDPLR
MAB_2689|M.abscessus_ATCC_1997 SLYLWHWPLLVYWLVYTGEARVTVAEGAAILGISLALAYLTNKYIETPLR
Mflv_3862|M.gilvum_PYR-GCK SLYLWHWPVLIFWLVHTGAPRVGILDGLGVLAVSLVLAYLTTRLIENPLR
: ******:*:::* ::* . . :* :* :* **:**.: :* ***
MSMEG_3187|M.smegmatis_MC2_155 TQ-----PGRAPVPAVPLRARLRRPTIVLGSIVALLGVALTATSFTWREH
Mvan_2787|M.vanbaalenii_PYR-1 SRGTV--AATAPVSKVALRKRLRRPTIVLGSTVALLGVALTATSFTWREH
TH_1799|M.thermoresistible__bu YRKPA--ADVVVPVKPPLRTRLRRPTMVLGSVVALLGVALTATSFTWREH
Mb1592c|M.bovis_AF2122/97 YRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTWREH
Rv1565c|M.tuberculosis_H37Rv YRAPAGVRSPAAVPPIPWRLRLRRPTIVLGSVVALLGVALTATSFTWREH
MMAR_2380|M.marinum_M YRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTWREH
MUL_1556|M.ulcerans_Agy99 YRAPSGTAAAVKTQAIPWQLRLRRPTIAMGSVVVLLGVTLTATSFTWREH
MLBr_01213|M.leprae_Br4923 YRTLANTEYPSPARAAAWQLRLHRSTIALGSMVVLLGVALTATSFTWRQH
MAV_3209|M.avium_104 HRGSARPAPRPAAPTVPWLARVPRPTMALGSAIVLLGVTLTATSFTWRQH
MAB_2689|M.abscessus_ATCC_1997 YP-------RVSPTSASLWTRLRRPTLALGTSIVLMAVALTATSFTWLEH
Mflv_3862|M.gilvum_PYR-GCK HPDSAG----DADRAARVWVWVRRPRFALGSAVVLLAVSLAVTSFAWREH
: *. :.:*: :.*:.*:*:.***:* :*
MSMEG_3187|M.smegmatis_MC2_155 VTVQRASGKELSGLSARDYPGARALIDHARVPKLPMRPTVLEAKNDLPIS
Mvan_2787|M.vanbaalenii_PYR-1 VTVLRANGKELSVLSLRGYPGALALVDNARVPELPMRPTVLEAKDDIPVS
TH_1799|M.thermoresistible__bu VSLQRASGKELSGLAARDYPGARALTEGARVAKLPMRPSVHEAREDLPIS
Mb1592c|M.bovis_AF2122/97 VIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDLPTS
Rv1565c|M.tuberculosis_H37Rv VIVQRAAGKELSGLSSRDYPGARALIDHVRVPKLRMRPTVLEVRHDLPTS
MMAR_2380|M.marinum_M VIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDFPAS
MUL_1556|M.ulcerans_Agy99 VIVERASGKELRGLSANDYPGARALTDHVRVPKLRMRPTVFEAKGDFPAS
MLBr_01213|M.leprae_Br4923 VIVLRATGKELSALNAQDYPGARALTAHARVPTLPMRPTVLEIKDDLPAS
MAV_3209|M.avium_104 VTVLRSAGKELSVLNPQDYPGARALTEHVRVPTLPMRPTVLEVKQDLPAS
MAB_2689|M.abscessus_ATCC_1997 VTVQRSNGKELSGLRPRDYPGAGALLYGDRVPDLPMRPTTLEASDDLPIT
Mflv_3862|M.gilvum_PYR-GCK VVMQRAKGHELAQLPERDYPGALALTNRIRVPQLPMRPTVLEARDDIPAT
* : *: *:** * ..**** ** **. * ***:. * *:* :
MSMEG_3187|M.smegmatis_MC2_155 TTEGCISDFANVGVINCTYGDKNATRTIALAGGSHAEHWITALDLLGQKH
Mvan_2787|M.vanbaalenii_PYR-1 TTEGCISDFDNVDIINCTYGDTAATRTIALAGGSHAEHWITALDLLGRAH
TH_1799|M.thermoresistible__bu TIDGCISDFDDVSVIKCTYGDESASRTIALAGGSHAEHWITALDLLGRKH
Mb1592c|M.bovis_AF2122/97 TKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLGRMH
Rv1565c|M.tuberculosis_H37Rv TKDGCISDFVNPAIINCTYGDVDAPRTIALAGGSHAEHWLTALDLLGRMH
MMAR_2380|M.marinum_M TRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLGRLH
MUL_1556|M.ulcerans_Agy99 TRDGCISDFVNPALVNCTYGDVDASRTIALAGGSHAEHWLPALDLLGRLH
MLBr_01213|M.leprae_Br4923 TRDGCISDFVNPAVVNCTYGDASANRTIALAGGSHAEHWLPALDVLGQLH
MAV_3209|M.avium_104 TRDGCISDFVNPAVVNCTYGDVTADRTIALAGGSHAEHWLPALDMLGKLH
MAB_2689|M.abscessus_ATCC_1997 TEQDCISDFRNRAVITCTYGNPHATRTIALAGGSHAEHWITALDILGRQH
Mflv_3862|M.gilvum_PYR-GCK TLDGCISDFVTVELINCIYGDPDAVRTIALAGGSHAEHWITALDTLGTRH
* :.***** ::.* **: * **************:.*** ** *
MSMEG_3187|M.smegmatis_MC2_155 GFKVVTYLKMGCPLTTEEVPLVSGDNRPYPKCHEWNQRVMAKLIADHPDY
Mvan_2787|M.vanbaalenii_PYR-1 HFKVVTYLKMGCPLTTEEKPLVMGDNRPYPKCREWNERVMPEIIADKPDF
TH_1799|M.thermoresistible__bu HFKVVTYLKMGCPLTTDEMPRVSGDGRPYPKCRDWNERVLPELIADRPDY
Mb1592c|M.bovis_AF2122/97 HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADHPDY
Rv1565c|M.tuberculosis_H37Rv HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCHQWVQAAMAKLVADHPDY
MMAR_2380|M.marinum_M HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDRPDY
MUL_1556|M.ulcerans_Agy99 HFKVVTYLKMGCPLSTEEVPLIMGNNAPYPQCREWVQKTMEKLVTDRPDY
MLBr_01213|M.leprae_Br4923 HFKVVTYLKMGCPLSTEHVPLIMGNNTPYPQCREWVQTTMTKLFSDRPDY
MAV_3209|M.avium_104 HFKVVTYLKMGCPLSTEQVPLIMGNNAPYPQCREWVQRTMTKLVTDRPDY
MAB_2689|M.abscessus_ATCC_1997 NFKITTYLKMGCPLSTEEVPRIAGSNDPYPDCKKWVDEVMSRIIQEHPDY
Mflv_3862|M.gilvum_PYR-GCK GFRVVTYLKMGCPLTTEMVPTIAISSEEYPQCHEWNQRAMDKLLTDRPDF
*::.*********:*: * : .. **.*:.* : .: .:. ::**:
MSMEG_3187|M.smegmatis_MC2_155 VFTTSTRPWNIK-PGDVMPSTYLGIWETFNENNIPVLAMRDTPWLTRNG-
Mvan_2787|M.vanbaalenii_PYR-1 VFTTSTRPWNIK-PGDVMPSSYVGIWQEFSDNNIPVLAMRDTPWMVRDG-
TH_1799|M.thermoresistible__bu IFTTSTRPWHTR-HGDRVPEGYLGIWEVLNEAGIPVLAMRDTPWLTKDGG
Mb1592c|M.bovis_AF2122/97 VFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVKDG-
Rv1565c|M.tuberculosis_H37Rv VFTTSTRPWNIK-PGDVMPATYVGIWQTFADNNIPVLAMRDTPWLVKDG-
MMAR_2380|M.marinum_M VFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVKDG-
MUL_1556|M.ulcerans_Agy99 VFTTSTRPWNIK-PGDVMPATYIGIWQTFADNNIPVLAVRDTPWMVKDG-
MLBr_01213|M.leprae_Br4923 VFTTSTRPWNTK-PGDVMPATYLGIWQALSDNNIPILAMRDTPWLVKNG-
MAV_3209|M.avium_104 VFTTTTRPWNIK-TGDVMPATYIGIWQTLSDNNIPILGMRDTPWLVKNG-
MAB_2689|M.abscessus_ATCC_1997 VFTTTTRPRSALGDGDVMPESYLGIWSAFDEAGIPVLGMRDTPWLIDKDG
Mflv_3862|M.gilvum_PYR-GCK VFTTTTRPNADE-PGDLVPEAYLGIWDTLSANGISILGMRDTPWLIRDG-
:***:*** ** :* *:***. : .*.:*.:*****: ..
MSMEG_3187|M.smegmatis_MC2_155 KPYFPADCLAD-GGDAVSCGVKRSKVLSDRNPTLDYLDRFPLMKPLDMSD
Mvan_2787|M.vanbaalenii_PYR-1 DPFFPADCLAD-GGDAISCGIERSEVLSDHNPTLDYVKRFPMLKPLDMSD
TH_1799|M.thermoresistible__bu RAKNPVDCLAD-GGDPVHCGMWRYEVLSDENPTLEWVETYPVLKPLDMSD
Mb1592c|M.bovis_AF2122/97 QPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLDMSD
Rv1565c|M.tuberculosis_H37Rv QPFIPADCLAK-GGNPQSCGIARSKVLVDRNPTLDFVARFPLLKPLDMSD
MMAR_2380|M.marinum_M QPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPIDMSD
MUL_1556|M.ulcerans_Agy99 QPFKPADCLAK-GGDAQSCGIKRSELLSDYNTTLDFVDRFPNLKPIDMSD
MLBr_01213|M.leprae_Br4923 QPSNPADCLAK-GGNAVSCGIKRSNVLADRNPTLNFVAQFSLLKPLDMSD
MAV_3209|M.avium_104 QPFDPADCLAKRGSTAQSCAIKRSDVLSERNQTLDFVGQFPQLKVLDMSD
MAB_2689|M.abscessus_ATCC_1997 TTYTAADCISA-GGTADSCAMDRNRALDDVNPTLAIANRFPLLKILDMTR
Mflv_3862|M.gilvum_PYR-GCK RLADPADCLSR-GGDSVSCGVKRSWALSETNPTLDLASYYPLLHPLDMSD
..**:: *. . *.: * * : * ** :. :: :**:
MSMEG_3187|M.smegmatis_MC2_155 AVCREDHCRVVEGNVLLYHDSHHLSATYMRTMTNELGRQMAAATGWW---
Mvan_2787|M.vanbaalenii_PYR-1 AVCRADHCRAVEGNVLLYHDAHHLSATYMRTMTDELGRQIGAATGWWSD-
TH_1799|M.thermoresistible__bu AVCRPDFCRVVEGNVLVYHDSHHFSSTYMRSMAPELGRQIAAVTGWWVPQ
Mb1592c|M.bovis_AF2122/97 AICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW---
Rv1565c|M.tuberculosis_H37Rv AICRTDTCRAVEGNVLVYRDSHHLTPTYMRTMTSELGRQIAANTDWW---
MMAR_2380|M.marinum_M AICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAITGWW---
MUL_1556|M.ulcerans_Agy99 AICRPDYCRAVEGNVLIYRDAHHMTPTYARTMAPELGRQIAAIIGWW---
MLBr_01213|M.leprae_Br4923 AICRPDICRAVEGNVLIYHGTHHLSPTYVRTLADELGRQIAENTGWW---
MAV_3209|M.avium_104 AICRADMCRPVEGNVLIYHGAHHMSPTYVRTMAPELGRQIAESTGWW---
MAB_2689|M.abscessus_ATCC_1997 AICRPDKCRVVEGNVLVYHDSHHISATYMRTMAKELGRQIALATGWWRPA
Mflv_3862|M.gilvum_PYR-GCK AVCDETDCRVTEGNILVYHDSHHVSATYMRTMTAELGRQIAAATGWW---
*:* ** .***:*:*:.:**.:.** *::: *****:. .**
MSMEG_3187|M.smegmatis_MC2_155 ----
Mvan_2787|M.vanbaalenii_PYR-1 ----
TH_1799|M.thermoresistible__bu ----
Mb1592c|M.bovis_AF2122/97 ----
Rv1565c|M.tuberculosis_H37Rv ----
MMAR_2380|M.marinum_M ----
MUL_1556|M.ulcerans_Agy99 ----
MLBr_01213|M.leprae_Br4923 ----
MAV_3209|M.avium_104 ----
MAB_2689|M.abscessus_ATCC_1997 QPGQ
Mflv_3862|M.gilvum_PYR-GCK ----