For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NMRVALFATCYNDLMWPETPKAVVRLLRRLGCEVEFPAEQTCCGQMFTNTGYAEEAIPAVRNFIATFAGY DAVVAPSGSCVGSVRHQHRDIAARAGCRAMQNEVDTLAPNVYELSEFLVDVLGVTDVGAYFPHRVTYHPT CHSLRMLRVGDRPLALLRAVRGIDLVELPGADQCCGFGGTFAVKNADTSVAMGADKARAVRETEAEVLVA GDNSCLAHIGGLLSRQRSGVRVMHLAEVLAAEGAS*
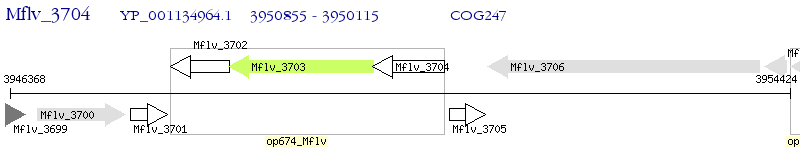
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3704 | - | - | 100% (246) | hypothetical protein Mflv_3704 |
| M. gilvum PYR-GCK | Mflv_0957 | - | 1e-06 | 24.49% (245) | FAD linked oxidase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0595 | - | 4e-88 | 66.12% (245) | glycolate oxidase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2709 | - | 1e-123 | 88.02% (242) | hypothetical protein Mvan_2709 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3704|M.gilvum_PYR-GCK MNMRVALFATCYNDLMWPETPKAVVRLLRRLGCEVEFPAEQTCCGQMFTN
Mvan_2709|M.vanbaalenii_PYR-1 MNTRVALFATCYNDLMWPETPKAVVRLLRRLGCHVEFPAAQTCCGQMFTN
MSMEG_0595|M.smegmatis_MC2_155 --MRIALFATCLADALFPPAAIATVQLLERLGHEVVFPSQQTCCGQMHIN
*:****** * ::* :. *.*:**.*** .* **: *******. *
Mflv_3704|M.gilvum_PYR-GCK TGYAEEAIPAVRNFIATFAG--YDAVVAPSGSCVGSVRHQHRDIAARAGC
Mvan_2709|M.vanbaalenii_PYR-1 TGYADEAIPAVRSFVAAFAG--YDAVVAPSGSCVGSVRHQHRDIAARARD
MSMEG_0595|M.smegmatis_MC2_155 TGYFKEAVAVVRNHVESFESARCDAVVAPSGSCVGSVRHQHAMVARRAGD
*** .**:..**..: :* . ****************** :* **
Mflv_3704|M.gilvum_PYR-GCK RAMQNEVDTLAPNVYELSEFLVDVLGVTDVGAYFPHRVTYHPTCHSLRML
Mvan_2709|M.vanbaalenii_PYR-1 PGLRSAVDAVAPNVYELSEFLVDVLGVTDVGAYFPHRVTYHPTCHSLRML
MSMEG_0595|M.smegmatis_MC2_155 EDLAVRAEKIAERTYELSEFLVDVLGVDDVGAYYPHRVTYHPTCHSLRML
: .: :* ..************* *****:****************
Mflv_3704|M.gilvum_PYR-GCK RVGDRPLALLRAVRGIDLVELPGADQCCGFGGTFAVKNADTSVAMGADKA
Mvan_2709|M.vanbaalenii_PYR-1 RVGDRPLRLLEAVRGIDLVALPRADQCCGFGGTFAMKNADTSVAMGFDKA
MSMEG_0595|M.smegmatis_MC2_155 GVGDKPLRLLRNVRGLTLVELPAAESCCGFGGTFALKNSDTSTAMLADKM
***:** **. ***: ** ** *:.*********:**:***.** **
Mflv_3704|M.gilvum_PYR-GCK RAVRETEAEVLVAGDNSCLAHIGGLLSRQRSGVRVMHLAEVLA-AEGAS-
Mvan_2709|M.vanbaalenii_PYR-1 RAARDTGAEVLVAGDNSCLAHIGGLLSRQRSGMRVMHLAEILASTEGDAA
MSMEG_0595|M.smegmatis_MC2_155 THILETGAEVCSAGDSSCLMHIGGGLSRLRSGVRTVHLAEILAGAEA---
:* *** ***.*** **** *** ***:*.:****:** :*.