For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAARQLSLSSSIAGLHGDERTVGSPLTAAELVAMRRTRLFGATGTVLMAIGALGAGARPVVQDPTFGVRL LNLPSRIATVSLTMTTTGAVMMALAWLMLGRFALGQRKMSRSQLDRTLMLWVLPLLIAPPMYSKDVYSYL AQSEISLQGNDPYEVGPATGLGLDHVFTLSVPSMWRETPAPYGPLFLWIGRGISRITGENIVEAVLFHRL VVLLGVALIVWATPRLARRCGVAEVSALWLGAANPLLFMHLVAGIHNEALMLGLMLAGTEFALRGIDAAA PLIPRPLVRPRDRHQWLRWYPLAMLFAGTVLITMSSQVKLPSLLALGFVAMALAYRWGGNVGAFLLASGA LGAVALAVVALIGWASGLGFGWLFTLGTANVVRSWMSPPTLLALGTGQVGILLGLGDHTTAVLALTRAIG VSLIAVIVLWLLFAVLRGRLHPVGGLGVALGATLLLFPVVQPWYVLWAIIPLAAWATRPRFRIAAIVVTL VVGIFGPTANGDRFALFQIALATAASFLILVMLVALTYRYLPWRPLPEDSREKSDAYAESP
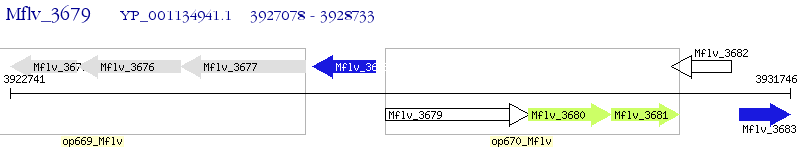
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3679 | - | - | 100% (551) | hypothetical protein Mflv_3679 |
| M. gilvum PYR-GCK | Mflv_2974 | - | 1e-61 | 30.81% (529) | hypothetical protein Mflv_2974 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1494c | - | 0.0 | 75.53% (564) | integral membrane protein |
| M. tuberculosis H37Rv | Rv1459c | - | 0.0 | 75.53% (564) | integral membrane protein |
| M. leprae Br4923 | MLBr_00591 | - | 0.0 | 73.52% (559) | hypothetical protein MLBr_00591 |
| M. abscessus ATCC 19977 | MAB_2751 | - | 0.0 | 73.08% (561) | hypothetical protein MAB_2751 |
| M. marinum M | MMAR_2264 | - | 0.0 | 77.72% (552) | integral membrane protein |
| M. avium 104 | MAV_3320 | - | 0.0 | 80.59% (541) | hypothetical protein MAV_3320 |
| M. smegmatis MC2 155 | MSMEG_3120 | - | 0.0 | 80.29% (558) | hypothetical protein MSMEG_3120 |
| M. thermoresistible (build 8) | TH_0709 | - | 1e-63 | 32.86% (496) | Possible conserved integral membrane protein |
| M. ulcerans Agy99 | MUL_1860 | - | 0.0 | 77.90% (552) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_2731 | - | 0.0 | 88.79% (562) | hypothetical protein Mvan_2731 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3679|M.gilvum_PYR-GCK ----------MAARQLSLSSSIAGLHGDERTVGSPLTAAELVAMRRTRLF
Mvan_2731|M.vanbaalenii_PYR-1 MGISARYAALVAARQHSLSSSIAGWHGDERTVGSPLTAAEVVAMRRTRLF
MSMEG_3120|M.smegmatis_MC2_155 ---------MMASRLSSFSSSIARWHGDEHAVASPLDDAEILSMRRTRLF
MMAR_2264|M.marinum_M ----------MAARHHTLSSSIASLHGDEQAVGSPLNAAELTALARTRLF
MUL_1860|M.ulcerans_Agy99 ----------MAARHHTLSSSIASLHGDEQAVGSPLNAAELTALARTRLF
Mb1494c|M.bovis_AF2122/97 ----------MAARHHTLSWSIASLHGDEQAVGAPLTTTELTALARTRLF
Rv1459c|M.tuberculosis_H37Rv ----------MAARHHTLSWSIASLHGDEQAVGAPLTTTELTALARTRLF
MLBr_00591|M.leprae_Br4923 ----------MAARHHTLSSSIASLHGDEQAVGSPLNDADRGALQRTRLF
MAV_3320|M.avium_104 ----------MAARHHTLSSSIASLHGDEQAVGSPLNDTELTALRRTRLF
MAB_2751|M.abscessus_ATCC_1997 ----------MSVRGQWLSSSLSHLHDDERSS-APLNEHELTAMNRTRLF
TH_0709|M.thermoresistible__bu ----------MTASTSSTDLPKSGLGG----QISRLNQFARSAAGRPALL
:: . . : . : * : *. *:
Mflv_3679|M.gilvum_PYR-GCK GATGTVLMAIGALGAGARPVVQDPTFGVRLLNLPS-RIATVSLTMTTTGA
Mvan_2731|M.vanbaalenii_PYR-1 GATGTVLMAIGALGAGARPVMQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MSMEG_3120|M.smegmatis_MC2_155 GATGTVLMAIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MMAR_2264|M.marinum_M GATGTVLMSIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MUL_1860|M.ulcerans_Agy99 GATGTVLMSIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
Mb1494c|M.bovis_AF2122/97 GATGTVLMAIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
Rv1459c|M.tuberculosis_H37Rv GATGTVLMAIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MLBr_00591|M.leprae_Br4923 GATGTILMAIGALGAGARPVIQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MAV_3320|M.avium_104 GASGTMLMAIGALGAGARPVVQDPTFGVRLLNLPS-RIQTVSLTMTTTGA
MAB_2751|M.abscessus_ATCC_1997 GATGTALMAIGALGAGARPVIQDPIFGVRLLNLPS-RIQTVSLTMTTTGA
TH_0709|M.thermoresistible__bu GGLGAVLITAGGLGAGSTRVHDPLLESLYLSWLRFGHGLVVSSTLLWTGV
*. *: *:: *.****: * : .: * * : .** *: **.
Mflv_3679|M.gilvum_PYR-GCK VMMALAWLMLGRFALG-----------------QRKMSRSQLDRTLMLWV
Mvan_2731|M.vanbaalenii_PYR-1 VMMTLAWLMLGRFTLG-----------------SRKMTRSQLDRTLLLWA
MSMEG_3120|M.smegmatis_MC2_155 VMMALAWLMLGRYTLG-----------------KRRMSRSQLDHTLMLWT
MMAR_2264|M.marinum_M VMMALAWLMLGRFALG-----------------KRRMSRGELDRTLVLWM
MUL_1860|M.ulcerans_Agy99 VMMALAWLMLGRFALG-----------------KRRMSRGELDRTLVLWM
Mb1494c|M.bovis_AF2122/97 VMMALAWLMLGRFTLG-----------------RRRMSRGELDRTLLLWM
Rv1459c|M.tuberculosis_H37Rv VMMALAWLMLGRFTLG-----------------RRRMSRGKLDRTLLLWM
MLBr_00591|M.leprae_Br4923 VMMTLAWLMLGRFALG-----------------RRQMSRGELDRTLLLWT
MAV_3320|M.avium_104 VMMALAWLMLGRFALG-----------------TRRMSRGDLDRTLLLWV
MAB_2751|M.abscessus_ATCC_1997 VMMTLAWLMLGRFAIGSLRNKGPDGRPDGPRGPVRRMSRGQLDRTLLLWL
TH_0709|M.thermoresistible__bu TLMLIAWLWLGRHVITG------------------RATEYGMIATTGFWL
.:* :*** ***..: : :. : * :*
Mflv_3679|M.gilvum_PYR-GCK LPLLIAPPMYSKDVYSYLAQSEISLQGNDPYEVGPATGLGLDHVFTLSVP
Mvan_2731|M.vanbaalenii_PYR-1 LPLLIAPPMYSKDVYSYLAQSEISLQGSDPYKVGPATGLGLDHVFTLSVP
MSMEG_3120|M.smegmatis_MC2_155 VPLLIAPPMYSRDVYSYLAQSEIAVLGLDPYRVGPATGLGLDHVFTLSVP
MMAR_2264|M.marinum_M LPLLIAPPMYSKDVYSYLAQSQISRDGLDPYRVGPASGLGLGHVFTLSVP
MUL_1860|M.ulcerans_Agy99 LPLLIAPPMYSKDVYSYLAQSQISRDGLDPYRVGPASGLGLDHVFTLSVP
Mb1494c|M.bovis_AF2122/97 LPLLIAPPMYSKDVYSYLAQSEIGRDGLDPYRVGPASGLGLGHVFTLSVP
Rv1459c|M.tuberculosis_H37Rv LPLLIAPPMYSKDVYSYLAQSEIGRDGLDPYRVGPASGLGLGHVFTLSVP
MLBr_00591|M.leprae_Br4923 LPLLIAPPMYSKDVYSYLAQSQISLQGLDPYRVGPASALGLGQVFTLSVP
MAV_3320|M.avium_104 LPLLIAPPMYSKDVYSYLAQSQISLEGLDPYRVGPASGLGLSHIFTLSVP
MAB_2751|M.abscessus_ATCC_1997 LPLLVAPPMYSKDVYSYLAQSQIARLGKDPYTEGPAAALGLDHVFTLSVP
TH_0709|M.thermoresistible__bu VPLLLSVPVFSRDTYSYLAQGALLRDGFDPYLVGPVENP---NPLLDDVS
:***:: *::*:*.******. : * *** **. : : .*.
Mflv_3679|M.gilvum_PYR-GCK SMWRETPAPYGPLFLWIGRGISRITGENIVEAVLFHRLVVLLGVALIVWA
Mvan_2731|M.vanbaalenii_PYR-1 SMWRETPAPYGPLFLWIGRGISRLTGENIVEAVLFHRLVVLLGVGLIVWA
MSMEG_3120|M.smegmatis_MC2_155 NLWRETPAPYGPLFLWIGQGISALTGENIVEAVMCHRLVVLIGVGLIVWA
MMAR_2264|M.marinum_M SLWRETPAPYGPLFLWIGRGISDLTGENIVAAVLFHRLVVLIGVALIVWA
MUL_1860|M.ulcerans_Agy99 SLWRETPAPYGPLFLWIGRGISDLTGENIVAAVLFHRLVVLIGVALIVWA
Mb1494c|M.bovis_AF2122/97 SLWRETPAPYGPLFLWIGRGISSLTGENIVAAVLCHRLVVLIGVTLIVWA
Rv1459c|M.tuberculosis_H37Rv SLWRETPAPYGPLFLWIGRGISSLTGENIVAAVLCHRLVVLIGVTLIVWA
MLBr_00591|M.leprae_Br4923 SLWRETPAPYGPLFLWIGRGISALTGDNIVAAVLCHRLMMLIGVGLVVWA
MAV_3320|M.avium_104 TLWRETPAPYGPLFLWIGRGISAITGENIVAAVLCHRLVELIGVGLIVWA
MAB_2751|M.abscessus_ATCC_1997 SLWRETPAPYGPLFLWIGKGISWLTGDDIVAGALCHRLVELLGVLLIIWA
TH_0709|M.thermoresistible__bu PIWTTTTAPYGPAFILVAKFVTMLVNDNVVAGTMLLRLCMLPGLALLIWA
:* *.***** *: :.: :: :..:::* ..: ** * *: *::**
Mflv_3679|M.gilvum_PYR-GCK TPRLARRCGVAEVSALWLGAANPLLFMHLVAGIHNEALMLGLMLAGTEFA
Mvan_2731|M.vanbaalenii_PYR-1 TPRLARRCGVAEVSALWLGPANPLLFMHLVAGIHNEALMLGLMLAGTEFA
MSMEG_3120|M.smegmatis_MC2_155 TPRLARRCGVAEVSALWLGPCNPLLFMHLVAGIHNEALMLGLMLAGTEFA
MMAR_2264|M.marinum_M TPRLAQRCGVAEVSALWLGAANPLLFMHLVAGIHNEALMLGLMLTGAEFA
MUL_1860|M.ulcerans_Agy99 TPRLAQRCGVAEVSALWLGAANPLLFMHLVAGIHNEALMLGLMLTGAEFA
Mb1494c|M.bovis_AF2122/97 TPRLAQRCGVAEVSALWLGAANPLLIMHLVAGIHNEALMLGLMLTGVEFA
Rv1459c|M.tuberculosis_H37Rv TPRLAQRCGVAEVSALWLGAANPLLIMHLVAGIHNEALMLGLMLTGVEFA
MLBr_00591|M.leprae_Br4923 TPQLARRCGVVEVSALWLGAANPLLIMHLVAGIHNEAPMLGLMLTGAEFA
MAV_3320|M.avium_104 TPRLARRCGVAEVSALWLGAANPLLIMHLVAGVHNEALMLGLMLAGAEFA
MAB_2751|M.abscessus_ATCC_1997 TPRLAQRCGVAEVSALWLGAANPLLLMHLIAGIHNEALMLGLMLAGTELA
TH_0709|M.thermoresistible__bu APRVARHIGANPAAALWICVLNPLVIIHLMGGVHNEMLMVGLMMAGIALT
:*::*:: *. .:***: ***:::**:.*:*** *:***::* ::
Mflv_3679|M.gilvum_PYR-GCK LRGIDAAAPLIPRPLVRPRD------------------------RHQWLR
Mvan_2731|M.vanbaalenii_PYR-1 LRGVDAAAPLLPRPLAWPRG------------------------RQQWQR
MSMEG_3120|M.smegmatis_MC2_155 LRGIDAAQPLLPRPLAWPSS------------------------RAQWQR
MMAR_2264|M.marinum_M LRGLDA---PRLIPLSGRPRPAPRRASSAE---------RSRAIKARAAA
MUL_1860|M.ulcerans_Agy99 LRGLDA---PRLIPLSGRPRPAPRRASSAE---------RSRAIKARAAA
Mb1494c|M.bovis_AF2122/97 LRGLDMANTPRPSPETWRLGPATIRASRRPELGASPRAGASRAVKP-RPE
Rv1459c|M.tuberculosis_H37Rv LRGLDMANTPRPSPETWRLGPATIRASRRPELGASPRAGASRAVKP-RPE
MLBr_00591|M.leprae_Br4923 LRGIDFSSTTRVIPTSWRGGGTGRQHSGTL---------TSQAIRF-GSN
MAV_3320|M.avium_104 LRGIDS---ARLLPHSLRPG----------------------------PD
MAB_2751|M.abscessus_ATCC_1997 MRGVDSTHSLAGVR-HWPRS------------------------HAQWAS
TH_0709|M.thermoresistible__bu LS--------------------------------------------RHPV
:
Mflv_3679|M.gilvum_PYR-GCK WYPLAMLFAGTVLITMSSQVKLPSLLALGFVAMALAYRWGGNVGAFLLAS
Mvan_2731|M.vanbaalenii_PYR-1 WYPLTMLITGAVLITLSSQVKLPSLLALGFVAMALACRWGGTVKAFVVAS
MSMEG_3120|M.smegmatis_MC2_155 WQPMAMLVLGAVLIAMSSQVKLPSLLALGFVAMALAWRWGGTVKAFVISC
MMAR_2264|M.marinum_M WEPLGMLAGGAILIALSSQVKLPSLLALGFITMALAYRYGGTIKALLVCG
MUL_1860|M.ulcerans_Agy99 WEPLGMLAGGAILIALSSQVKLPSLLALGFITMALAYRYGGTIKALLVCG
Mb1494c|M.bovis_AF2122/97 WGPLAMLLAGSILITLSSQVKLPSLLAMGFVTTVLAYRWGGNLRALLLAA
Rv1459c|M.tuberculosis_H37Rv WGPLAMLLAGSILITLSSQVKLPSLLAMGFVTTVLAYRWGGNLRALLLAA
MLBr_00591|M.leprae_Br4923 WEPLGMLLAGSILIVLSSQVKLPSLLALGFVAMALACRLGSTPQALLMVS
MAV_3320|M.avium_104 WEPLGMLLAGAVLITLSSQVKLPSLLALGFVTMALAYRCGGNLRALLLAG
MAB_2751|M.abscessus_ATCC_1997 WAPLGNLLAGTLLITASSQVKLPSLLALGFVGMALARRWGGGVRGLFQAG
TH_0709|M.thermoresistible__bu WG--VASIAVAVAVKATAGLALP-FMVWVWMRQLREQRSLSPAAAFAIAT
* :: : :: : ** ::. :: * . .:
Mflv_3679|M.gilvum_PYR-GCK GALGAVALAVVALIGWASGLGFGWLFTLGTANVVRSWMSPPTLLALGTGQ
Mvan_2731|M.vanbaalenii_PYR-1 GSLGAVALAVMALIGWASGLGFGWLFTLGTANVVRSWMSPPTLLALGTGQ
MSMEG_3120|M.smegmatis_MC2_155 TSLGAISLAVMAVIGWASGLGFGWLFTLGTANVVRSWMSPPTLIALGTGQ
MMAR_2264|M.marinum_M SAMSALALSVMAVVGWASGLGFGWMFTLGTANVVRSWMSPPTLLALGTGQ
MUL_1860|M.ulcerans_Agy99 SAMSALALSVMAVVGWASGLGFGWMFTLGTANVVRSWMSPPTLLALGTGQ
Mb1494c|M.bovis_AF2122/97 AVMASLTLAIMAILGWASGLGFGWINTLGTANVVRSWMSPPTLLALGTGH
Rv1459c|M.tuberculosis_H37Rv AVMASLTLAIMAILGWASGLGFGWINTLGTANVVRSWMSPPTLLALGTGH
MLBr_00591|M.leprae_Br4923 LWMAGLSLAVMGIVGWASGLGFGWIYTLGTANVVRSWMSPPTLLALGTGH
MAV_3320|M.avium_104 GGMAALSLAVMALVGWASGLGFGWIYTLGTANVVRSWMSPPTLLALGTGQ
MAB_2751|M.abscessus_ATCC_1997 TLLGAVTGACTVLIGWASGLGFGWIYTLGTANAVRSWMSPPTLVALGTGQ
TH_0709|M.thermoresistible__bu AASLAIFAAVFAVLSWVAGVGLGWLTALAGSVKIINWLTVPTAVANLTNA
.: : ::.*.:*:*:**: :*. : : .*:: ** :* *.
Mflv_3679|M.gilvum_PYR-GCK VGILLGLGDHTTAVLALTRAIGVSLIAVIVLWLLFAVLRGRLHPVGGLGV
Mvan_2731|M.vanbaalenii_PYR-1 VGILLGLGDHTTAVLALTRAMGVTLIAVIVLWLLFAVLRGRLHPVGGLGV
MSMEG_3120|M.smegmatis_MC2_155 VGILLGLGDHTTAVLGLTRAIGVFMISILVSWLLFAVLRGRLHPVGGLGV
MMAR_2264|M.marinum_M VGILLRLGDHTTAVLSLTRAMGVLIIMVMVSWLLLAVFRGRLHPIGGLGV
MUL_1860|M.ulcerans_Agy99 VGILLRLGDHTTAVLSLTRAMGVLIIIVMVSWLLLAVFRGRLHPIGGLGV
Mb1494c|M.bovis_AF2122/97 VGILLGLGDHTTAVLSLTRAIGVLIITVMVCWLLLAVLRGRLHPIGGLGV
Rv1459c|M.tuberculosis_H37Rv VGILLGLGDHTTAVLSLTRAIGVLIITVMVCWLLLAVLRGRLHPIGGLGV
MLBr_00591|M.leprae_Br4923 VGILLGLGDHTTAVLSLTRGIGVLIIMVMACWLLLAVFRGRLHPIGGLGV
MAV_3320|M.avium_104 VGILLGLGDHTTAVLGLTRAIGVLIITVMVGWLLLAVFRGRLHPIGGLGV
MAB_2751|M.abscessus_ATCC_1997 VGILLGLGDHTTAILSLTRGIGIGIIAIAVSWLLFAVLRGRLHPVGGLGV
TH_0709|M.thermoresistible__bu IGGMF-LPVNFYAVLEVTRIIGIAIIVVTLPVVWWRFRHDDRGALTGIAV
:* :: * : *:* :** :*: :* : : . :. .: *:.*
Mflv_3679|M.gilvum_PYR-GCK ALGATLLLFPVVQPWYVLWAIIPLAAWAT-RPRFRIAAIVVTLVVGIFGP
Mvan_2731|M.vanbaalenii_PYR-1 ALGATLLLFPVVQPWYVLWAIIPLAAWAT-RPRFRAAAIVVTLVVGIFGP
MSMEG_3120|M.smegmatis_MC2_155 ALGGTVLLFPVVQPWYLLWAIIPLAAWAT-RPGFRGATIAITLIVGIFGP
MMAR_2264|M.marinum_M ALGLTVLLFPVVQPWYLLWAIIPMAAWAT-RTGFRVATIIITLIVGIFGP
MUL_1860|M.ulcerans_Agy99 ALGLTVLLFPVVQPWYLLWAIIPMAAWAT-RTGFRVATIIITLIVGIFGP
Mb1494c|M.bovis_AF2122/97 ALAVTVLLFPVVQPWYLLWAIIPLAAWAT-RPGFRVAAILATLIVGIFGP
Rv1459c|M.tuberculosis_H37Rv ALAVTVLLFPVVQPWYLLWAIIPLAAWAT-RPGFRVAAILATLIVGIFGP
MLBr_00591|M.leprae_Br4923 ALGVTVLLFPVVQPWYLLWAIIPLAAWAT-RTGFRVAAITVTIVVGVFGP
MAV_3320|M.avium_104 ALGVTVLLFPVVQPWYLLWAIIPLAAWAT-RTGFRVAAIVISLVVGIFGP
MAB_2751|M.abscessus_ATCC_1997 ALGITVLLFPVVQPWYLLWAIIPLAAWAT-RPAFRGTAIGITLLVGIFGP
TH_0709|M.thermoresistible__bu AMLVVVLFVPAALPWYYTWPLAVLSALAQSRPAIALIAGFSTWIMVIFRP
*: .:*:.*.. *** *.: ::* * *. : : : :: :* *
Mflv_3679|M.gilvum_PYR-GCK TANGDRFALFQIALATAASFLILVMLVALTYRYLPWR-------------
Mvan_2731|M.vanbaalenii_PYR-1 TANGDRFALFQIALATVASTAILLILLALTYRRLPWRVRPGEPPD-----
MSMEG_3120|M.smegmatis_MC2_155 TANGDRFTLFQIVMATLASAVTVLLLIALTYRRLPWRPAPEPP-------
MMAR_2264|M.marinum_M TANGDRFALFQIVDATLASAIIVLVLMALTYHRLPWRALPVESPPPAEVT
MUL_1860|M.ulcerans_Agy99 TANGDRFALFQIVDATLASAIIVLVLMALTYHRLPWRALPVESPPPAEVT
Mb1494c|M.bovis_AF2122/97 TANGDRFALFQIVDATAASAIIVILLIALTYTRLPWRPLAAE--------
Rv1459c|M.tuberculosis_H37Rv TANGDRFALFQIVDATAASAIIVILLIALTYTRLPWRPLAAE--------
MLBr_00591|M.leprae_Br4923 TANGDRFALFQIVDATVASTIIVLVLIALTYTRLPWRPLQMVPAKRSELD
MAV_3320|M.avium_104 TANGDRFALFQIVDATLASTVIVAVLIALTYTRLPWRPLPAP--QRAPDG
MAB_2751|M.abscessus_ATCC_1997 TANGDRFALFQIIDATAASAIIALLLIAITHDYLPWRRLPNGHQEAPLTL
TH_0709|M.thermoresistible__bu DGAHGMYSWTHVLIATGCAVAAWWWLYRTEST------------------
. . :: :: ** .: *
Mflv_3679|M.gilvum_PYR-GCK -----------------------------PLPEDSREKS---DAYAESP-
Mvan_2731|M.vanbaalenii_PYR-1 --------------------------EAAPPPAPPHAKP---DAYAESP-
MSMEG_3120|M.smegmatis_MC2_155 ----------------------------ARPPEQPAPAD---DAYAESP-
MMAR_2264|M.marinum_M QPAEASQQAEPNQQGESDQPETADPAAKPPATRRPTSTT---DAYADST-
MUL_1860|M.ulcerans_Agy99 QPAEASQQAEPNQQGESNQPETADPAAKPPATRRPTSTT---DAYADST-
Mb1494c|M.bovis_AF2122/97 QVVTAAESA-----------------SKTPATRRPTAAP---DAYADST-
Rv1459c|M.tuberculosis_H37Rv QVVTAAESA-----------------SKTPATRRPTAAP---DAYADST-
MLBr_00591|M.leprae_Br4923 QQLVRVVAP-----------------DPETSQNSASPATRLPTAYANLT-
MAV_3320|M.avium_104 QPVIDPVAP-----------------ARSPSTPRPTPAH---DAYADST-
MAB_2751|M.abscessus_ATCC_1997 TDRDATIDQDS-------------TTSADAVGDQAIRDGRGSDHSVGQPG
TH_0709|M.thermoresistible__bu -------------------------------PAEPIPQS---GGSAGESG
. . .
Mflv_3679|M.gilvum_PYR-GCK -
Mvan_2731|M.vanbaalenii_PYR-1 -
MSMEG_3120|M.smegmatis_MC2_155 -
MMAR_2264|M.marinum_M -
MUL_1860|M.ulcerans_Agy99 -
Mb1494c|M.bovis_AF2122/97 -
Rv1459c|M.tuberculosis_H37Rv -
MLBr_00591|M.leprae_Br4923 -
MAV_3320|M.avium_104 -
MAB_2751|M.abscessus_ATCC_1997 P
TH_0709|M.thermoresistible__bu R