For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AEFSMRYIVVDVDEAVNFYTRHLGFELVMKPGPGFAMLRREGVRLLVNAPEGGGGAGQRLSGGELPQPGG WNRFQVRVDDLAAVVDALRRAGVPFRGEVIEGRGGRQALAVDPSGNLVELFQVNPG*
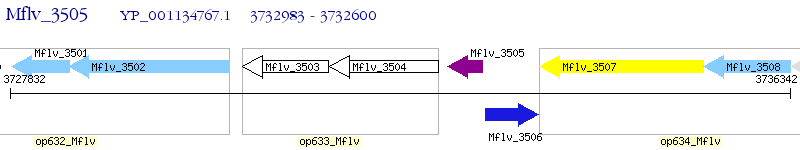
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3505 | - | - | 100% (127) | glyoxalase/bleomycin resistance protein/dioxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0592 | TB27.3 | 3e-05 | 31.20% (125) | hypothetical protein Mb0592 |
| M. tuberculosis H37Rv | Rv0577 | TB27.3 | 3e-05 | 31.20% (125) | hypothetical protein Rv0577 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3357 | - | 3e-06 | 32.35% (136) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3286 | - | 2e-56 | 80.16% (126) | glyoxalase/bleomycin resistance protein/dioxygenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3505|M.gilvum_PYR-GCK MAEFS---------MRYIVVDVDEAVNFYTRHLGFELVMKPGPG------
Mvan_3286|M.vanbaalenii_PYR-1 MTDFS---------VRYIVDDVDDAVDFYTRHLGFELVMKPGPG------
Mb0592|M.bovis_AF2122/97 MPKRSEYRQGTPNWVDLQTTDQSAAKKFYTSLFGWGYDDNPVPGGGGVYS
Rv0577|M.tuberculosis_H37Rv MPKRSEYRQGTPNWVDLQTTDQSAAKKFYTSLFGWGYDDNPVPGGGGVYS
TH_3357|M.thermoresistible__bu MSDRIR-----LTHFGLCVRDIGRAEQFYCNALGFEAIGRMTVDDDATAT
*.. . . * . * .** :*: . .
Mflv_3505|M.gilvum_PYR-GCK FAMLRREGVRLLVNAPEG---------------------------GGG--
Mvan_3286|M.vanbaalenii_PYR-1 FAMLRRGGLRLLLNVPEG---------------------------GGG--
Mb0592|M.bovis_AF2122/97 MATLNGEAVAAIAPMPPGAPEGMPPIWNTYIAVDDVDAVVDKVVPGGGQV
Rv0577|M.tuberculosis_H37Rv MATLNGEAVAAIAPMPPGAPEGMPPIWNTYIAVDDVDAVVDKVVPGGGQV
TH_3357|M.thermoresistible__bu LLDIPGLLLELVYLQRDGFRLELLG-------------------------
: : : : *
Mflv_3505|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_3286|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb0592|M.bovis_AF2122/97 MMPAFDIGDAGRMSFITDPTGAAVGLWQANRHIGATLVNETGTLIWNELL
Rv0577|M.tuberculosis_H37Rv MMPAFDIGDAGRMSFITDPTGAAVGLWQANRHIGATLVNETGTLIWNELL
TH_3357|M.thermoresistible__bu --------------------------------------------------
Mflv_3505|M.gilvum_PYR-GCK ----------------------------------AGQRLSGGELPQ----
Mvan_3286|M.vanbaalenii_PYR-1 ----------------------------------AGQRLSGGQLPQ----
Mb0592|M.bovis_AF2122/97 TDKPDLALAFYEAVVGLTHSSMEIAAGQNYRVLKAGDAEVGGCMEPPMPG
Rv0577|M.tuberculosis_H37Rv TDKPDLALAFYEAVVGLTHSSMEIAAGQNYRVLKAGDAEVGGCMEPPMPG
TH_3357|M.thermoresistible__bu ---------------------------------YARPATVGEQGPRPMN-
* *
Mflv_3505|M.gilvum_PYR-GCK -PGGWNRFQVRVDDLAAVVDALRRAG--VPFRGEVIEGRGGRQALAVDPS
Mvan_3286|M.vanbaalenii_PYR-1 -PGGWNRFQLQVGDLDAVVEALHSGG--VAFRGEVVEGRGGRQALALDPS
Mb0592|M.bovis_AF2122/97 VPNHWHVYFAVDDADATAAKAAAAGG--QVIAEPADIPSVGRFAVLSDPQ
Rv0577|M.tuberculosis_H37Rv VPNHWHVYFAVDDADATAAKAAAAGG--QVIAEPADIPSVGRFAVLSDPQ
TH_3357|M.thermoresistible__bu -AVGFTHLSFRVDDADRVMDAIVAHGGRTLPERTVAFSGGNRGLMATDPE
. : . . .* * . .* : **.
Mflv_3505|M.gilvum_PYR-GCK GNLVELFQVNPG-
Mvan_3286|M.vanbaalenii_PYR-1 GNPVELFQAYPA-
Mb0592|M.bovis_AF2122/97 GAIFSVLKPAPQQ
Rv0577|M.tuberculosis_H37Rv GAIFSVLKPAPQQ
TH_3357|M.thermoresistible__bu GNLLELIERTADR
* ..::: .