For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRTDSVGYRDENLGVGAVLTTAAFFCVALVGTLAKVGGDHTSTGVLLLFQNAVGVLFLLPVVFRADRRML RTDRIWLHVLRAATGTACWYALFFAITQIPLANATLLTYSAPLWMPLIAWAVTRHRVAAPTWLGAGIGFV GILMVLQPQGHGFSAGELSALAGALMLAVAMMSVRWLGATEPVLRILFYYFLLSTVMSLPIAIADWQPVG AAGWPWLAGLGFAQLFSQALIVLAYRYASAEKLGPFIYTVIVFTAVIDWIVWDHRPTLSTYIGMALVVGG GLFAMRARSPAAVSG
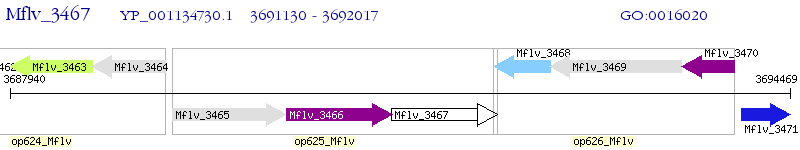
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3467 | - | - | 100% (295) | hypothetical protein Mflv_3467 |
| M. gilvum PYR-GCK | Mflv_3133 | - | 6e-10 | 27.30% (293) | hypothetical protein Mflv_3133 |
| M. gilvum PYR-GCK | Mflv_4964 | - | 2e-05 | 28.83% (163) | hypothetical protein Mflv_4964 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0677c | - | 7e-06 | 25.82% (244) | hypothetical protein MAB_0677c |
| M. marinum M | MMAR_4118 | - | 2e-05 | 25.20% (250) | membrane permease |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1505 | - | 4e-05 | 24.91% (281) | integral membrane protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3984 | - | 3e-05 | 25.20% (250) | membrane permease |
| M. vanbaalenii PYR-1 | Mvan_3392 | - | 4e-09 | 26.09% (253) | hypothetical protein Mvan_3392 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4118|M.marinum_M ---MASRTPLPVLMILGSCISLQLGAAIATPLLAHFGAGL----TTGVRL
MUL_3984|M.ulcerans_Agy99 ---MASRTPLPVLMILGSCVSLQLGAAIATPLLAHFGAGL----TTGVRL
Mvan_3392|M.vanbaalenii_PYR-1 -------------MAVGSMVCVQLGLAIAVTLIEDIGVEG----AAWLRL
MSMEG_1505|M.smegmatis_MC2_155 MTEGPRRPAGAAVLVVVASISQEVGAAFAVGLFATLGTLG----AVFTRF
Mflv_3467|M.gilvum_PYR-GCK ---MRTDSVGYRDENLGVGAVLTTAAFFCVALVGTLAKVGGDHTSTGVLL
MAB_0677c|M.abscessus_ATCC_199 ------------MTTRHRLLGLTVAVLWGVNFVAIHAGLE-----HFPPL
. . :. . :
MMAR_4118|M.marinum_M LFAATLLMAIHRPRAFGWDRQQWRSAALFAVALAGMN-----GFFFAAIA
MUL_3984|M.ulcerans_Agy99 LFAATLLMAIHRPRAFGWDRQQWRSAALFAVALAGMN-----GFFFAAIA
Mvan_3392|M.vanbaalenii_PYR-1 AWAGILFLVIVRPRRKAFTRNSFLMCVLLGVVTAAIT-----LLFMAALA
MSMEG_1505|M.smegmatis_MC2_155 AVAGVILCAAVRPRLRGVSGRAWLSALALATTLVVMN-----FSFYCAIA
Mflv_3467|M.gilvum_PYR-GCK LFQNAVGVLFLLPVVFRADRRMLRTDRIWLHVLRAATGTACWYALFFAIT
MAB_0677c|M.abscessus_ATCC_199 FFAGLRFAVLALPVILFVPWPRVPVRWLLLYGLGFGT-IQFAFLFLAMAN
* . :
MMAR_4118|M.marinum_M RIPLGVAVTIEFAG----PLMLAAAFSRRPRDLGCVMAAAAAIVVLSWDG
MUL_3984|M.ulcerans_Agy99 RIPLGVAVTIEFAG----PLMLAAAFSRRPRDLGCVMAAAAAIVVLSWDG
Mvan_3392|M.vanbaalenii_PYR-1 RIPLGTASALEFLG----PLGVAVARGRGQG--RWIWPGLAAVGVLSLTQ
MSMEG_1505|M.smegmatis_MC2_155 RIPLGIAVTIEVLG----PLILSVAASTRRVAWLWAVLAFAGVAMLGLTR
Mflv_3467|M.gilvum_PYR-GCK QIPLANATLLTYSAPLWMPLIAWAVTRHRVAAPTWLGAGIGFVGILMVLQ
MAB_0677c|M.abscessus_ATCC_199 GMPTGLASLVLQASAPFTAILGVVLLGERMSAVQVTGISLAVAGMALIAA
:* . * : . .: . . . :
MMAR_4118|M.marinum_M SKS-AALSPDLAGVAFALVAACFWALYILAGKRITGQQTG-HGALSVSMF
MUL_3984|M.ulcerans_Agy99 SKS-AALSPDLAGVAFALVAACFWALYILAGKRITGQQTG-HGALSVSKF
Mvan_3392|M.vanbaalenii_PYR-1 PWS-AAVDP--VGVGYALCAAACWAGYILLTQKVGDEVAG-INGLAVSMP
MSMEG_1505|M.smegmatis_MC2_155 HDD-GRLDA--AGFGFAAVAGASWAGYILASSRTAAQFRR-IDGLALATG
Mflv_3467|M.gilvum_PYR-GCK PQG-HGFSA---GELSALAGALMLAVAMMSVRWLGATEPV-LRILFYYFL
MAB_0677c|M.abscessus_ATCC_199 DRATHGASAAVVPVVVTLLGALGWAFGNIASRKAMEHNTDPHTPLRLTLW
.. : .. * : *
MMAR_4118|M.marinum_M IAAIVVLPFGIPAVPTLTGQ--------------PEKLIPLLGVAVLSSM
MUL_3984|M.ulcerans_Agy99 IAAIVVLPFGIPAVPTLTGQ--------------PEKLIPLLGVAVLSSM
Mvan_3392|M.vanbaalenii_PYR-1 VAGLVATVTVGPAVIERMT---------------PQILLIGIGLAILLPV
MSMEG_1505|M.smegmatis_MC2_155 LGALALTPFALLLVDFAGALR-------------FDVIVLAVSVSVLSSL
Mflv_3467|M.gilvum_PYR-GCK LSTVMSLPIAIADWQPVGAAG-------------WPWLAGLG----FAQL
MAB_0677c|M.abscessus_ATCC_199 MSVVPPIPLFALSAFLEGPDAGWKALTSIGTHSGLIALAGLVYIVVLATI
:. : : : :
MMAR_4118|M.marinum_M LPYSLEFAAMRRFSAHTFGVLLSLEPVVAGLAGWILLGQTLSWVRGLAMV
MUL_3984|M.ulcerans_Agy99 LPYSLEFAAMRRFSAHTFGVLLSLEPVVAGLAGWILLGQTLSWVRGLAMV
Mvan_3392|M.vanbaalenii_PYR-1 VPFALELLALRRLTAAAFGTLMALEPAFAMLLGFLILNQAPGAAGVIGIL
MSMEG_1505|M.smegmatis_MC2_155 VPYSLELLSLRHLHPSVFAVLTSLSPVVAALAGLMVLGQQIGTLGYLGVA
Mflv_3467|M.gilvum_PYR-GCK FSQALIVLAYRYASAEKLGPFIYTVIVFTAVIDWIVWDHRPTLSTYIGMA
MAB_0677c|M.abscessus_ATCC_199 VGLGLWTYLMGRYPASRVAPFSLLVPIVGITTSWLALGERPTALELVGAA
. .* . .. : . . : .. :.
MMAR_4118|M.marinum_M VVVAASIICTLSQPSPASEPSSAKERTPTRDRAQGSAVGRPLARVRNGLQ
MUL_3984|M.ulcerans_Agy99 VVVPASIICTLSQPSPASEPSSAKERTPTRDRAQGSAVGRPLARVRNGLQ
Mvan_3392|M.vanbaalenii_PYR-1 FVVAAGVGAARSGARAAPVPLEVG--------------------------
MSMEG_1505|M.smegmatis_MC2_155 LVTAASIGAVRTAHTGPAEPLA----------------------------
Mflv_3467|M.gilvum_PYR-GCK LVVGGGLFAMRARSPAAVSG------------------------------
MAB_0677c|M.abscessus_ATCC_199 VVITGCLIGMTARPAARAPQHTDTPERGPRELSACAAPAGH---------
.* . : :
MMAR_4118|M.marinum_M TSQRREPVAGRDGLPSPRAEWLDADVSLGDSGRFGDKIARISHRANPVDG
MUL_3984|M.ulcerans_Agy99 TAQRREAVAGREGVPSPAAEWLDADVSLGDSGRFGDKIARISHRANPVDG
Mvan_3392|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1505|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3467|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0677c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4118|M.marinum_M GRGEKAPIRSAAQVGPHFGATGSATNHSATDVPRWADEHHSPATHVTSQS
MUL_3984|M.ulcerans_Agy99 GRGEKAPIRSAAQVGPHFGATGSATNHSATDVPRWADEHHSPATHVTSQS
Mvan_3392|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1505|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_3467|M.gilvum_PYR-GCK --------------------------------------------------
MAB_0677c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_4118|M.marinum_M ILRRRVRHRHHPARPGPEGAGIQQ
MUL_3984|M.ulcerans_Agy99 ILRRRVRHRHHPARPGPEGAGIQQ
Mvan_3392|M.vanbaalenii_PYR-1 ------------------------
MSMEG_1505|M.smegmatis_MC2_155 ------------------------
Mflv_3467|M.gilvum_PYR-GCK ------------------------
MAB_0677c|M.abscessus_ATCC_199 ------------------------