For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDGLPIAVATPLRCGAAALMIVLAAAGCSSAPAHETSAHAAATAPALTGSLDEWRAAICRDDVTTVSGT RHTVEGSTCVPHDGDGVVNFDRFESEPSMSSMLSWQPSVYVAHAVVDDRPLAIWTPSGEASDLEPLTRYG FTLAAYESPTLSRPTAELPATVPSGDIEVIPPNAYGYVVVQTAGGDTQCLVDTTYVGCETEGTNWQQHLD GSGPYHGVRINADGTGSWVDGNLGAAAPQTLGDRTYRALGWTIVSSPAGMRFTNDRTGHGAIVSAAKVQT F
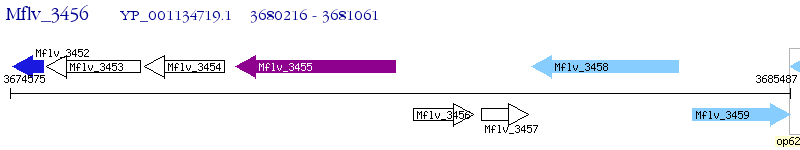
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3456 | - | - | 100% (281) | hypothetical protein Mflv_3456 |
| M. gilvum PYR-GCK | Mflv_0264 | - | 3e-25 | 44.37% (151) | putative lipoprotein LpqJ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0351c | lpqJ | 2e-26 | 38.80% (183) | lipoprotein LpqJ |
| M. tuberculosis H37Rv | Rv0344c | lpqJ | 2e-26 | 38.80% (183) | lipoprotein LpqJ |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1827c | - | 1e-08 | 33.82% (136) | bacteriophage protein |
| M. marinum M | MMAR_0620 | lpqJ | 7e-23 | 37.87% (169) | lipoprotein, LpqJ |
| M. avium 104 | MAV_4811 | - | 2e-23 | 46.92% (130) | LpqJ protein |
| M. smegmatis MC2 155 | MSMEG_0704 | - | 2e-21 | 42.54% (134) | LpqJ protein |
| M. thermoresistible (build 8) | TH_1024 | lpqJ | 1e-23 | 44.78% (134) | PROBABLE LIPOPROTEIN LPQJ |
| M. ulcerans Agy99 | MUL_0585 | lpqJ | 4e-23 | 38.10% (168) | lipoprotein, LpqJ |
| M. vanbaalenii PYR-1 | Mvan_3213 | - | 7e-84 | 74.86% (183) | LpqJ |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3456|M.gilvum_PYR-GCK MSDGLPIAVATPLRCGAAALMIVLAAAGCSSAPAHETSAHAAATAPALTG
Mvan_3213|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0704|M.smegmatis_MC2_155 --------------------------------------------------
TH_1024|M.thermoresistible__bu --------------------------------------------------
Mb0351c|M.bovis_AF2122/97 --------------------------------------------------
Rv0344c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_0620|M.marinum_M --------------------------------------------------
MUL_0585|M.ulcerans_Agy99 --------------------------------------------------
MAV_4811|M.avium_104 --------------------------------------------------
MAB_1827c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3456|M.gilvum_PYR-GCK SLDEWRAAICRDDVTTVSGTRHTVEGSTCVPHDGDGVVNFDRFESEPSMS
Mvan_3213|M.vanbaalenii_PYR-1 ------------------------------------------------MD
MSMEG_0704|M.smegmatis_MC2_155 ------------------------------------------MKVRLLLG
TH_1024|M.thermoresistible__bu ------------------------------VRRFVGPVGAVVLGVTALIA
Mb0351c|M.bovis_AF2122/97 -------------------------------MRLSLIAR--------GMA
Rv0344c|M.tuberculosis_H37Rv -------------------------------MRLSLIAR--------GMA
MMAR_0620|M.marinum_M -------------------------------MRLPQKTRW-------GLL
MUL_0585|M.ulcerans_Agy99 -------------------------------MRLPQKTRW-------GLL
MAV_4811|M.avium_104 -------------------------------MRLSENARRSLAGGALLLV
MAB_1827c|M.abscessus_ATCC_199 MSAEDDEDEQEPTRASPTALAETGVVEITPAAWSDSDEIDDIDPYEDPVR
:
Mflv_3456|M.gilvum_PYR-GCK SMLSWQPSVYVAHAVVDDRPLAIWTPSGEASDLEPLTRYGFTLAAYESPT
Mvan_3213|M.vanbaalenii_PYR-1 SVLSWQPSTHTAETVVDGRPLAIWTPSGEAADLEPLKQFGFTVASYQSAS
MSMEG_0704|M.smegmatis_MC2_155 CLLSVGGLLVGCHSAIEGKPVTK-AITPIEPNFPTPRPTRSSP--APSST
TH_1024|M.thermoresistible__bu VLIAVG--VAGCSRTTEGSPVGT-GPTGEEPTFPTPRPTRTTP--PTTTP
Mb0351c|M.bovis_AF2122/97 ALLAATALVAGCNTTIDGRPVASPGSGPTEPTFPTPRPTTAPP--GTTAP
Rv0344c|M.tuberculosis_H37Rv ALLAATALVAGCNTTIDGRPVASPGSGPTEPTFPTPRPTTAPP--GTTAP
MMAR_0620|M.marinum_M ALSAAAVLAAGCDATIDGKPIATPGAGPTEPSFPTRRPTTSPPPPTSTSP
MUL_0585|M.ulcerans_Agy99 ALSAAAVLAAGCDATIDGKPIATPGAGPTEPSFPTRRPTTSPP--TSTSP
MAV_4811|M.avium_104 VLPGGGGLVSGCSTVIGGRPVASPGAGPTEPSFPTPR--STPP--SATAA
MAB_1827c|M.abscessus_ATCC_199 RNWLISGAIFAATAAVAGLAAGGAFVFLRQERGKTPVTTSTVLIASPPAP
. . . . . ...
Mflv_3456|M.gilvum_PYR-GCK LSRPTAELPATVPSGDIEVIPPNAYGYVVVQTAGGDTQCLVDTTYVGCET
Mvan_3213|M.vanbaalenii_PYR-1 FVRATADVPAALPSGDIETIPPNEYGYVVVQTAAGDTQCIVEATFIGCET
MSMEG_0704|M.smegmatis_MC2_155 VAAPP----SAKPPTGATPLQP-QNGYVFIETKSGRTRCQLSKQDVGCES
TH_1024|M.thermoresistible__bu PPDTG----TATPSPRGEVLRP-QNGYVFIETKSGQTRCQLSADEVGCEA
Mb0351c|M.bovis_AF2122/97 TLPTT--PVSPTAPAGAIPLPPDSNGYVFIETKSGMTRCQINRDSVGCEA
Rv0344c|M.tuberculosis_H37Rv TLPTT--PVSPTAPAGAIPLPPDSNGYVFIETKSGMTRCQINRDSVGCEA
MMAR_0620|M.marinum_M SQPTS--PASPTSPAGAIPLPPDDNGYVFIETKSGQTRCQINHDSVGCEA
MUL_0585|M.ulcerans_Agy99 SQPTS--PASPTSPAGAIPLPPDDNGYVFIETKSGQTRCQINHDSVGCEA
MAV_4811|M.avium_104 PAPTALPPPSPSAPAGAIPLPPDQNGYVFIETKSGSTRCQINKDTVGCEA
MAB_1827c|M.abscessus_ATCC_199 TSAVVLPPPPTQSRPPTPIELSATGDSVYVSTKSGKTACQVTVNTVSCIV
.. . . . * :.* .* * * : :.*
Mflv_3456|M.gilvum_PYR-GCK EGTNWQQHLDGSGPYHGVRINADGTGSWVDGNLGAAAPQTLGD-RTYRAL
Mvan_3213|M.vanbaalenii_PYR-1 EGTNWQQHVDGSGPYHGVRINTDGTGSWVDGNLGAVAPTTLGA-KTYRAL
MSMEG_0704|M.smegmatis_MC2_155 DFANAPV-VDGL-PANGVQLSASGKMRWIVGNLGAIPAVTLDY-KRYSAL
TH_1024|M.thermoresistible__bu PFHDPPE-VDGV-PANGVNITADGAVRWVVGNLGAIPAVTLDY-RTYSAL
Mb0351c|M.bovis_AF2122/97 PFTNSPL-RDGE-HANGIHITAGGSVQWVLGNLGAIPTVSIDY-RTYEAQ
Rv0344c|M.tuberculosis_H37Rv PFTNSPL-RDGE-HANGIHITAGGSVQWVLGNLGAIPTVSIDY-RTYEAQ
MMAR_0620|M.marinum_M PFTNSPI-KDGE-HANGVHITSAGNVQWVLGNLGAIPTVTIDY-KTYDAQ
MUL_0585|M.ulcerans_Agy99 PFTNSPI-KDGE-HANGVHITSAGNVQWVLGNLGAIPTVTIDY-KTYDAQ
MAV_4811|M.avium_104 PFTNSPL-QDGE-HANGVSINAGGKVQWVLGNLGAIPTVKLDY-RTYSAQ
MAB_1827c|M.abscessus_ATCC_199 RFVGRTPIRYGV-PTNVVMITSGGIMDWTVGDGGQLQTHTLNYGTLYHAL
. * : : :.: * * *: * . .:. * *
Mflv_3456|M.gilvum_PYR-GCK GWTIVSSPAGMRFTNDRTGHGAIVSAAKVQTF
Mvan_3213|M.vanbaalenii_PYR-1 GWTIASTPAGMRFTNDKTGHGALVSVQNVQPF
MSMEG_0704|M.smegmatis_MC2_155 GWTIDASSDGTRFTNDGTGHGMVVAVEGVQVF
TH_1024|M.thermoresistible__bu GWTIEATRDGTRFTNDRTGHGMFVAVEGVETF
Mb0351c|M.bovis_AF2122/97 GWTIDATTDGTRFTNNRTGHGMFVSIEKVDTF
Rv0344c|M.tuberculosis_H37Rv GWTIDATTDGTRFTNNRTGHGMFVSIEKVDTF
MMAR_0620|M.marinum_M GWIITATTDGTRFQNNGTGHGMFVSIDKVETF
MUL_0585|M.ulcerans_Agy99 GWIITATTDGTRFQNNGTGHGMFVSIDKVETF
MAV_4811|M.avium_104 GWTIVANADGTRFTNDQTKHGMFVSIEKVNTF
MAB_1827c|M.abscessus_ATCC_199 GWTITPTSEGTTFTNDATGRGMSVNVDGARAF
** * .. * * *: * :* * . *