For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDERDFSDVLRSAALRVTRPRMAVLSAVGQHPHADTEVIIRAARQELPDISRQTVYDALNALTATGLVRR IQPAGSLARYESRVGDNHHHIVCRSCGVIADVDCAVGDTPCLTASDDLGFEVDEAEVIYWGLCPECSTTD APAH*
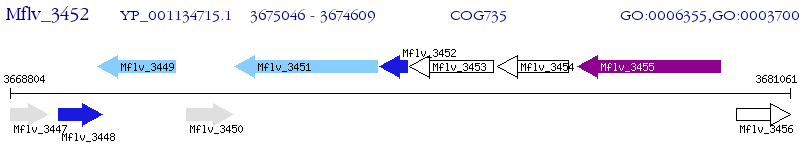
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3452 | - | - | 100% (145) | ferric uptake regulator family protein |
| M. gilvum PYR-GCK | Mflv_2717 | - | 2e-06 | 24.41% (127) | ferric uptake regulator family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1944c | furA | 8e-59 | 75.74% (136) | ferric uptake regulation protein furA (fur) |
| M. tuberculosis H37Rv | Rv1909c | furA | 8e-59 | 75.74% (136) | ferric uptake regulation protein furA (fur) |
| M. leprae Br4923 | MLBr_00824 | furB | 5e-07 | 24.03% (129) | putative ferric uptake regulatory protein |
| M. abscessus ATCC 19977 | MAB_2471c | - | 2e-53 | 67.15% (137) | ferric uptake regulation protein FurA |
| M. marinum M | MMAR_2915 | furA | 4e-54 | 73.08% (130) | ferric uptake regulation protein FurA |
| M. avium 104 | MAV_2752 | - | 2e-52 | 73.85% (130) | transcriptional regulator, Fur family protein |
| M. smegmatis MC2 155 | MSMEG_6383 | - | 1e-54 | 67.39% (138) | transcription regulator FurA |
| M. thermoresistible (build 8) | TH_0864 | furA | 2e-45 | 67.77% (121) | FERRIC UPTAKE REGULATION PROTEIN FURA (FUR) |
| M. ulcerans Agy99 | MUL_2189 | furA | 1e-54 | 73.85% (130) | ferric uptake regulation protein FurA |
| M. vanbaalenii PYR-1 | Mvan_3209 | - | 1e-68 | 82.52% (143) | ferric uptake regulator family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3452|M.gilvum_PYR-GCK ---MTDERDFSDVLRSAALRVTRPRMAVLSAVG-QHPHADTEVIIRAARQ
Mvan_3209|M.vanbaalenii_PYR-1 MEPMTEEPDYSAVLRSAALRVTRPRMAVLHAVG-RHPHADTELIINATRE
MMAR_2915|M.marinum_M ---------------MADLRVTRPRLAVLEAMG-DHPHADTETIYSAVRE
MUL_2189|M.ulcerans_Agy99 ---------------MADLRVTRPRLAVLEAVG-DHPHADTETIYSAVRE
MAV_2752|M.avium_104 ---------------MADLRVTRPRVAVLEVVD-ANPHADTETIFSAVRM
MAB_2471c|M.abscessus_ATCC_199 --MAFTSDEYAALLRTADLRVTRPRVAVLEAVE-ANPHADTETVFGVVRT
TH_0864|M.thermoresistible__bu ---------------------------VLEAVH-DHPHADTESIYLAVRA
Mb1944c|M.bovis_AF2122/97 MSSVSSIPDYAEQLRTADLRVTRPRVAVLEAVN-AHPHADTETIFGAVRF
Rv1909c|M.tuberculosis_H37Rv MSSVSSIPDYAEQLRTADLRVTRPRVAVLEAVN-AHPHADTETIFGAVRF
MSMEG_6383|M.smegmatis_MC2_155 ---MTHTTDFEQLLRSAGMRVTRPRIAVLRSVY-AHPHADTDTVIRAVRE
MLBr_00824|M.leprae_Br4923 ------------MMTGASVRSTRQRAAISTLLETVDDFRSAQELHNELRR
: : . . .:: : *
Mflv_3452|M.gilvum_PYR-GCK ELPDISRQTVYDALNALTATGLVRRIQPAGSLARYESRVGDNHHHIVCRS
Mvan_3209|M.vanbaalenii_PYR-1 LVPDVSRQTVYDALNALAATGLVRRIQPAGSVARYETRVGDNHHHVVCRN
MMAR_2915|M.marinum_M ILPDVSRQAVYDVLGALTSVGLVRRIQPSGSVARYESRVGDNHHHVVCRS
MUL_2189|M.ulcerans_Agy99 ILPDVSRQAVYDVLGALTSVGLVRRIQPSGSVARYESRVGDNHHHVVCRS
MAV_2752|M.avium_104 ALPDVSRQAVYDVLNALTAVGLVRRIQPLGMVARYESRVGDNHHHVVCRS
MAB_2471c|M.abscessus_ATCC_199 GLPEVSRQAVYDVLAALTSAGLIRKIQPSGSVARYESRVGDNHHHVVCRS
TH_0864|M.thermoresistible__bu KLPAVSRQAVYDVLHALTATGLARRIQPPGSVARYEARVGDNHHHLVCRS
Mb1944c|M.bovis_AF2122/97 ALPDVSRQAVYDVLHALTAAGLVRKIQPSGSVARYESRVGDNHHHIVCRS
Rv1909c|M.tuberculosis_H37Rv ALPDVSRQAVYDVLHALTAAGLVRKIQPSGSVARYESRVGDNHHHIVCRS
MSMEG_6383|M.smegmatis_MC2_155 ALPEVSHQAVYDSLHALAAARLVRRIQPSGSVARYESRVGDNHHHVVCRA
MLBr_00824|M.leprae_Br4923 RGNNISLTTVYRTLQSMAEAGMVDTLRTNTGESVYRRCSRQHHHHLVCRG
:* :** * ::: . : ::. : *. ::***:***
Mflv_3452|M.gilvum_PYR-GCK CGVIADVDCAVGDTPCLTASDD----LGFEVDEAEVIYWGLCPECSTTDA
Mvan_3209|M.vanbaalenii_PYR-1 CGVIADVDCAVGEAPCLTASDD----LGFEVDEAEVIYWGICPACSTAPS
MMAR_2915|M.marinum_M CGAIADIDCAVGEAPCLAPSDDSNVLDGFVVDEAEVIFWGMCPDCSTAMP
MUL_2189|M.ulcerans_Agy99 CGAIADIDCAVGEAPCLAPSDDSNVLDGFVVDEAEVIFWGMCPDCSTAMP
MAV_2752|M.avium_104 CGTIADVDCAVGEAPCLTPSDDDNVLDGFVLDEAEVIYWGLCADCSTAGS
MAB_2471c|M.abscessus_ATCC_199 CGAIADVDCAVGAAPCLTPSD----YNGFILEEAEVIYWGLCPDCASTEA
TH_0864|M.thermoresistible__bu CGSVADIDCAVGAAPCLTPADPGGVLDGFLLDEAEVIYWGVCPDCARTEP
Mb1944c|M.bovis_AF2122/97 CGVIADVDCAVGEAPCLTASD----HNGFLLDEAEVIYWGLCPDCSISDT
Rv1909c|M.tuberculosis_H37Rv CGVIADVDCAVGEAPCLTASD----HNGFLLDEAEVIYWGLCPDCSISDT
MSMEG_6383|M.smegmatis_MC2_155 CGDIADVDCAVGEAPCLTGSDH----HGYAIDEAEVIYWGTCPACAQDPD
MLBr_00824|M.leprae_Br4923 CGSTIEVGDHEVEAWAAEVAAK----HGFSDVSHTIEIFGTCSECRS---
** ::. : . : *: . : :* *. *
Mflv_3452|M.gilvum_PYR-GCK PAH--------
Mvan_3209|M.vanbaalenii_PYR-1 PQ---------
MMAR_2915|M.marinum_M RSQP-------
MUL_2189|M.ulcerans_Agy99 RSQP-------
MAV_2752|M.avium_104 -----------
MAB_2471c|M.abscessus_ATCC_199 APRISGSHT--
TH_0864|M.thermoresistible__bu I----------
Mb1944c|M.bovis_AF2122/97 SRSHP------
Rv1909c|M.tuberculosis_H37Rv SRSHP------
MSMEG_6383|M.smegmatis_MC2_155 RAKARTSRSHP
MLBr_00824|M.leprae_Br4923 -----------