For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RGTVDTSGVTAGARPLIATAASEEQRVTDKDFNSGGDSDEVTVETTSVFRADFLNELDAPPAAGGDSAVS GVEGLPAGSALLVVKRGPNAGSRFLLDQPTTSAGRHPDSDIFLDDVTVSRRHAEFRLESGEFQVVDVGSL NGTYVNREPVDSAVLANGDEVQIGKFRLVFLTGPKGDDGANG*
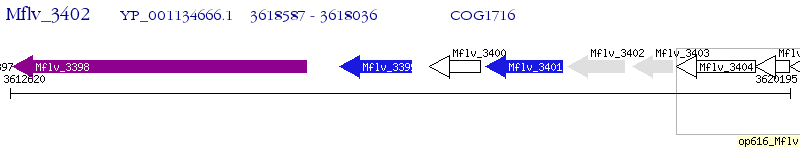
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3402 | - | - | 100% (183) | FHA domain-containing protein |
| M. gilvum PYR-GCK | Mflv_4882 | - | 2e-11 | 40.87% (115) | ABC transporter related |
| M. gilvum PYR-GCK | Mflv_0807 | - | 2e-09 | 42.35% (85) | FHA domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1858 | cfp17 | 4e-69 | 84.38% (160) | hypothetical protein Mb1858 |
| M. tuberculosis H37Rv | Rv1827 | cfp17 | 4e-69 | 84.38% (160) | hypothetical protein Rv1827 |
| M. leprae Br4923 | MLBr_02076 | - | 8e-68 | 85.71% (154) | hypothetical protein MLBr_02076 |
| M. abscessus ATCC 19977 | MAB_2403 | - | 9e-68 | 85.90% (156) | hypothetical protein MAB_2403 |
| M. marinum M | MMAR_2704 | - | 2e-70 | 85.62% (160) | hypothetical protein MMAR_2704 |
| M. avium 104 | MAV_2888 | - | 7e-71 | 87.26% (157) | forkhead-associated protein |
| M. smegmatis MC2 155 | MSMEG_3647 | - | 7e-71 | 92.91% (141) | forkhead-associated protein |
| M. thermoresistible (build 8) | TH_1126 | - | 5e-68 | 90.00% (140) | CONSERVED HYPOTHETICAL PROTEIN CFP17 |
| M. ulcerans Agy99 | MUL_3046 | - | 8e-70 | 85.00% (160) | hypothetical protein MUL_3046 |
| M. vanbaalenii PYR-1 | Mvan_3135 | - | 1e-81 | 97.39% (153) | FHA domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2704|M.marinum_M -------------------------MTDMDAARDRDQTSDEVTVETTSVF
MUL_3046|M.ulcerans_Agy99 -------------------------MTDMDAARDRDQTSDEVTVETTSVF
MLBr_02076|M.leprae_Br4923 -------------------------MTDMDSGRQEDQTSDEVTVETTSVF
MAV_2888|M.avium_104 -------------------------MTDMDSGSQ-DQNGDEVTVETTSVF
Mb1858|M.bovis_AF2122/97 -------------------------MTDMNPDIEKDQTSDEVTVETTSVF
Rv1827|M.tuberculosis_H37Rv -------------------------MTDMNPDIEKDQTSDEVTVETTSVF
Mflv_3402|M.gilvum_PYR-GCK MRGTVDTSGVTAGARPLIATAASEEQRVTDKDFNSGGDSDEVTVETTSVF
Mvan_3135|M.vanbaalenii_PYR-1 ---------------------------MTDKDFNSGTDSDEVTVETTSVF
MSMEG_3647|M.smegmatis_MC2_155 -----------------------------------------MTVETTSVF
TH_1126|M.thermoresistible__bu -------------------------------------------VETTSVF
MAB_2403|M.abscessus_ATCC_1997 --------------------------MVTDND--KDDTSGEVTAETTSVF
.******
MMAR_2704|M.marinum_M RADFLNELDAPTQSGTESSVSGVEGLPAGSALLVVKRGPNAGSRFLLDQP
MUL_3046|M.ulcerans_Agy99 RADFLNELDAPTQSGTESSVSGVEGLPAGSALLVVKRGPNAGSRFLLDQP
MLBr_02076|M.leprae_Br4923 RADFLNELDAPAQAGAESVVSGVEGLLAGSALLVVKRGPNAGSRFLLDQA
MAV_2888|M.avium_104 RADFLNELDAPAQAGTESAVSGVEGLPAGSALLVVKRGPNAGSRFLLDQA
Mb1858|M.bovis_AF2122/97 RADFLSELDAPAQAGTESAVSGVEGLPPGSALLVVKRGPNAGSRFLLDQA
Rv1827|M.tuberculosis_H37Rv RADFLSELDAPAQAGTESAVSGVEGLPPGSALLVVKRGPNAGSRFLLDQA
Mflv_3402|M.gilvum_PYR-GCK RADFLNELDAPPAAGGDSAVSGVEGLPAGSALLVVKRGPNAGSRFLLDQP
Mvan_3135|M.vanbaalenii_PYR-1 RADFLNELDAPPAAGGESAVSGVEGLPVGSALLVVKRGPNAGSRFLLDQP
MSMEG_3647|M.smegmatis_MC2_155 RADFLNELDAPAAAGTEGAVSGVEGLPSGSALLVVKRGPNAGSRFLLDQP
TH_1126|M.thermoresistible__bu RADFLSELDAPAQAGTEGAVSGVEGLPPGSALLVVKRGPNAGSRFLLDQP
MAB_2403|M.abscessus_ATCC_1997 RADFATELEAPAQAGAD--VSGVEGLPAGSALLVVKRGPNAGSRFLLDQA
**** .**:**. :* : ******* *********************.
MMAR_2704|M.marinum_M ITSAGRHPDSDIFLDDVTVSRRHAEFRLEGNEFSVVDVGSLNGTYVNREP
MUL_3046|M.ulcerans_Agy99 ITSAGRHPDSDIFLDDVTVSRRHAEFRLEGNEFSVVDVGSLNGTYVNRGP
MLBr_02076|M.leprae_Br4923 ITSAGRHPDSDIFLDDVTVSRRHAEFRLEGNEFHVVDVGSLNGTYVNREP
MAV_2888|M.avium_104 TTSAGRHPDSDIFLDDVTVSRRHAEFRLENNEFSVVDVGSLNGTYVNREP
Mb1858|M.bovis_AF2122/97 ITSAGRHPDSDIFLDDVTVSRRHAEFRLENNEFNVVDVGSLNGTYVNREP
Rv1827|M.tuberculosis_H37Rv ITSAGRHPDSDIFLDDVTVSRRHAEFRLENNEFNVVDVGSLNGTYVNREP
Mflv_3402|M.gilvum_PYR-GCK TTSAGRHPDSDIFLDDVTVSRRHAEFRLESGEFQVVDVGSLNGTYVNREP
Mvan_3135|M.vanbaalenii_PYR-1 TTSAGRHPDSDIFLDDVTVSRRHAEFRLESGEFQVVDVGSLNGTYVNREP
MSMEG_3647|M.smegmatis_MC2_155 TTSAGRHPDSDIFLDDVTVSRRHAEFRLEGGEFQVVDVGSLNGTYVNREP
TH_1126|M.thermoresistible__bu TTSAGRHPDSDIFLDDVTVSRRHAEFRLEGGEFQVVDVGSLNGTYVNREP
MAB_2403|M.abscessus_ATCC_1997 TTSAGRHPDSDIFLDDVTVSRRHAEFRLDSAEFQVVDVGSLNGTYVNREP
***************************:. ** ************** *
MMAR_2704|M.marinum_M VDSAVLANGDEVQIGKFRLVFLTGPKQGDDDGGTGGQ
MUL_3046|M.ulcerans_Agy99 VDSAVLANGDEVQIGKFRLVFLTGPKQGDDDGGTGGQ
MLBr_02076|M.leprae_Br4923 VDSAVLANGDEVQIGKFRLVFLIGSKQDDDTGRTAGK
MAV_2888|M.avium_104 VDSAVLANGDEVQIGKFRLVFLTGPKQGEDGG-SSG-
Mb1858|M.bovis_AF2122/97 VDSAVLANGDEVQIGKFRLVFLTGPKQGEDDGSTGGP
Rv1827|M.tuberculosis_H37Rv VDSAVLANGDEVQIGKFRLVFLTGPKQGEDDGSTGGP
Mflv_3402|M.gilvum_PYR-GCK VDSAVLANGDEVQIGKFRLVFLTGPK--GDDGANG--
Mvan_3135|M.vanbaalenii_PYR-1 VDSAVLANGDEVQIGKFRLVFLTGPK--GADGGASD-
MSMEG_3647|M.smegmatis_MC2_155 VDSAVLANGDEVQIGKFRLVFLTGPK--SDDSGSNA-
TH_1126|M.thermoresistible__bu VDSAVLANGDEVQIGKFRLVFLTGPK--SEESGPGS-
MAB_2403|M.abscessus_ATCC_1997 VDSAVLANGDEVQIGKFRLVFLTGPKSAGDDSASGGQ
********************** *.* .