For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTWPDRDTSAFPSALPGAFDASSAMSRGVVNLSSFSDFLRTQAPHLLPVSGSPGVTPSDAVPHGTTIVAL KFPGGVVMAGDRRATQGHMIASRDVQKVYITDDYTVTGIAGTAAIAVEFARLYAVELEHYEKLEGVALTF PGKVNRLATMVRGNLGAALQGFVALPLLAGYDLDDPNPEGAGRIVSFDAAGGHNLEEEGYQSVGSGSIFA KSSMKKLYSQVTDADSALRVAIEALYDAADDDSATGGPDLVRGIFPTAVVITADGAVEIAEQQIADMCRE IIQNRSRADTFGPDHAPVRGDEL
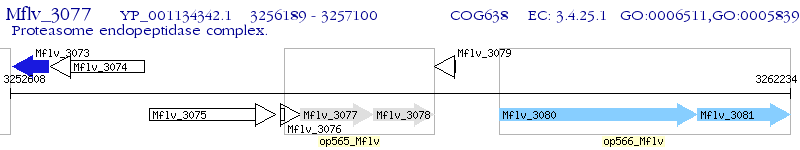
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3077 | - | - | 100% (303) | 20S proteasome, A and B subunits |
| M. gilvum PYR-GCK | Mflv_2743 | - | 7e-09 | 25.00% (220) | hypothetical protein Mflv_2743 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2134c | prcB | 1e-116 | 72.05% (297) | proteasome (beta subunit) PrcB |
| M. tuberculosis H37Rv | Rv2110c | prcB | 1e-116 | 72.05% (297) | proteasome (beta subunit) PrcB |
| M. leprae Br4923 | MLBr_01322 | prcB | 1e-112 | 73.26% (288) | proteasome beta-type subunit 2 |
| M. abscessus ATCC 19977 | MAB_2172 | - | 2e-96 | 68.22% (258) | proteasome (beta subunit) PrcB |
| M. marinum M | MMAR_3085 | prcB | 1e-117 | 73.22% (295) | proteasome (beta subunit) PrcB |
| M. avium 104 | MAV_2404 | - | 1e-121 | 75.33% (304) | PcrB protein |
| M. smegmatis MC2 155 | MSMEG_3895 | - | 1e-122 | 81.78% (269) | proteasome beta subunit |
| M. thermoresistible (build 8) | TH_0505 | prcB | 1e-124 | 83.83% (266) | proteasome (beta subunit) PrcB |
| M. ulcerans Agy99 | MUL_2331 | prcB | 1e-117 | 73.22% (295) | proteasome (beta subunit) PrcB |
| M. vanbaalenii PYR-1 | Mvan_3449 | - | 1e-143 | 84.64% (306) | 20S proteasome, A and B subunits |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2134c|M.bovis_AF2122/97 MTWPLPDRLSINSLSGT-----------PAVDLSSFTDFLRRQAPELLPA
Rv2110c|M.tuberculosis_H37Rv MTWPLPDRLSINSLSGT-----------PAVDLSSFTDFLRRQAPELLPA
MMAR_3085|M.marinum_M MTWPLPDRLSINSAISG-----------SAVDLSSFAEFLRRQAPELLPA
MUL_2331|M.ulcerans_Agy99 MTWPLPDRLSINSAISG-----------SAVDLSSFAEFLRRQAPELLPA
MLBr_01322|M.leprae_Br4923 MTRSFPDRLPTNLAFPG----------ISVINQSSFVDLLRRQAPELLPV
MAV_2404|M.avium_104 MTWQFPDRLSTNSALPGP-----AFSGQPAVDLSSFADFLRRQAPELLPA
Mflv_3077|M.gilvum_PYR-GCK MTWPDRDTSAFPSALPGAFDAS-SAMSRGVVNLSSFSDFLRTQAPHLLPV
Mvan_3449|M.vanbaalenii_PYR-1 MTWPNRDQPAFPSSSSGAFSALPQGVSHGGVNLSSFSDFLRHQAPHLLPG
MSMEG_3895|M.smegmatis_MC2_155 ------------------------------MDLSSFSELLSRQAPELLPV
TH_0505|M.thermoresistible__bu ------------------------------VHLSSFSDLLRRQAPHLLPG
MAB_2172|M.abscessus_ATCC_1997 MIWRDDAIASQPAGQHFR-----------NSHLSSFSEYLRAQAPELLPA
. *** : * ***.***
Mb2134c|M.bovis_AF2122/97 ---SISG-GAP---LAGGDAQLPHGTTIVALKYPGGVVMAGDRRSTQGNM
Rv2110c|M.tuberculosis_H37Rv ---SISG-GAP---LAGGDAQLPHGTTIVALKYPGGVVMAGDRRSTQGNM
MMAR_3085|M.marinum_M ---SIKH-GGG---AVG--DQLPHATTIVALKYPGGVLIAGDRRSTQGNM
MUL_2331|M.ulcerans_Agy99 ---SIKH-GGG---AVG--DQLPHATTIVALKYPGGVLIAGDRRSTQGNM
MLBr_01322|M.leprae_Br4923 ---SLGGGQSG-----GGQ-QLSHGTTIVVLKYPGGVVIAGDRRSTQGNM
MAV_2404|M.avium_104 ---ALGGAQSGPASSSGGTGQLPHGTTIVALKYPGGVVLAGDRRSTQGNM
Mflv_3077|M.gilvum_PYR-GCK ---SGSPGVT-------PSDAVPHGTTIVALKFPGGVVMAGDRRATQGHM
Mvan_3449|M.vanbaalenii_PYR-1 ---APGPGPTGL-----PTDAVPHGTTIVALKCPGGVVMAGDRRATQGNM
MSMEG_3895|M.smegmatis_MC2_155 NRVAYGTTPVG------PTDAVPHGTTIVALKYPGGVLIAGDRRSTQGNM
TH_0505|M.thermoresistible__bu ---AGDPPPVA------AGDALPHGTTIVALKYPGGVVIAGDRRSTQGNM
MAB_2172|M.abscessus_ATCC_1997 LG-KASADVIG---------NLPHGTTIVALTYRGGVLIAGDRRATQGNY
:.*.****.*. ***::*****:***:
Mb2134c|M.bovis_AF2122/97 ISGRDVRKVYITDDYTATGIAGTAAVAVEFARLYAVELEHYEKLEGVPLT
Rv2110c|M.tuberculosis_H37Rv ISGRDVRKVYITDDYTATGIAGTAAVAVEFARLYAVELEHYEKLEGVPLT
MMAR_3085|M.marinum_M IAGRDVRKVYITDDYTATGIAGTAAIAVEFARLYAVELEHYEKLEGVPLT
MUL_2331|M.ulcerans_Agy99 IAGRDVRKVYITDDYTATGIAGTAAIAVEFARLYAVELEHYEKLEGVPLT
MLBr_01322|M.leprae_Br4923 IAGRDVRKVYITDDYTATGIAGIAAVAVEFARLYAVELEHYEKLEGVPLT
MAV_2404|M.avium_104 IAGRDVKKVYITDDYTATGIAGTAAIAVEFARLYAVELEHYEKLEGVPLT
Mflv_3077|M.gilvum_PYR-GCK IASRDVQKVYITDDYTVTGIAGTAAIAVEFARLYAVELEHYEKLEGVALT
Mvan_3449|M.vanbaalenii_PYR-1 IASRDVQKVYITDDYTVTGIAGTAAIAVEFARLYAVELEHYEKLEGVALT
MSMEG_3895|M.smegmatis_MC2_155 IAGRDVQKVYITDDYTATGIAGTAAIAVEFARLYAVELEHYEKLEGVPLT
TH_0505|M.thermoresistible__bu IAGRDVQKVYITDDYTATGIAGTAAIAVEFARLYAVELEHYEKLEGVPLT
MAB_2172|M.abscessus_ATCC_1997 IANRDIDKVQITDNYSATGIAGTAAIAVEFARLYAVELEHYEKLEGVQLT
*:.**: ** ***:*:.***** **:********************* **
Mb2134c|M.bovis_AF2122/97 FAGKINRLAIMVRGNLAAAMQGLLALPLLAGYDIHASDPQSAGRIVSFDA
Rv2110c|M.tuberculosis_H37Rv FAGKINRLAIMVRGNLAAAMQGLLALPLLAGYDIHASDPQSAGRIVSFDA
MMAR_3085|M.marinum_M FAGKVNRLAIMVRGNLAAAMQGLVALPLLASYDIHASDPRSAGRIVSFDA
MUL_2331|M.ulcerans_Agy99 FAGKVNRLAIMVRGNLAAAMQGLVALPLLASYDIHASDPRSAGRIVSFDA
MLBr_01322|M.leprae_Br4923 FAGKVNRLAIMVRSNLTAAMQGLLALPLLAGYDIHAPDPQSAGRIVSFDA
MAV_2404|M.avium_104 FAGKVNRLAIMVRGNLAAAMQGLVALPLLAGYDIDARDPEGAGRIVSFDA
Mflv_3077|M.gilvum_PYR-GCK FPGKVNRLATMVRGNLGAALQGFVALPLLAGYDLDDPNPEGAGRIVSFDA
Mvan_3449|M.vanbaalenii_PYR-1 FPGKVNRLATMVRGNLGAALQGFVALPLLAGYDLDDPNPEAAGRIVSFDA
MSMEG_3895|M.smegmatis_MC2_155 FRGKVNRLAIMVRGNLGAALQGFVALPLLVGYDLDDPHPEGAGRIVSFDA
TH_0505|M.thermoresistible__bu FNGKVNRLAIMVRSNLGAALQGFVALPLLVGYDLDDPNPEAAGRIVSFDA
MAB_2172|M.abscessus_ATCC_1997 FNGKANRLSALVRGNLAAAMQGLVAVPLLVGYDLDDPNGETAGRIVSFDV
* ** ***: :**.** **:**::*:***..**:. . . ********.
Mb2134c|M.bovis_AF2122/97 AGGWNIEEEGYQAVGSGSLFAKSSMKKLYSQVTDGDSGLRVAVEALYDAA
Rv2110c|M.tuberculosis_H37Rv AGGWNIEEEGYQAVGSGSLFAKSSMKKLYSQVTDGDSGLRVAVEALYDAA
MMAR_3085|M.marinum_M AGGWNIEEEGYQAVGSGSIFAKSSIKKLYAQVTDADSALRVAVEALYDAA
MUL_2331|M.ulcerans_Agy99 AGGWNIEEEGYQAVGSGSIFAKSSIKKLYAQVTDADSALRVAVEALYDAA
MLBr_01322|M.leprae_Br4923 AGGWNIEEEGYQSVGSGSIFAKSSIKKLYSQVSDADSALRVAIEALYDAA
MAV_2404|M.avium_104 AGGWNLEEEGYQSVGSGSIFAKSSMKKLYSQVVDADSAVRVAIEALYDAA
Mflv_3077|M.gilvum_PYR-GCK AGGHNLEEEGYQSVGSGSIFAKSSMKKLYSQVTDADSALRVAIEALYDAA
Mvan_3449|M.vanbaalenii_PYR-1 AGGWNFEEEGYQSVGSGSIFAKSSMKKLYSQVTDAESALRVAVEALYDAA
MSMEG_3895|M.smegmatis_MC2_155 AGGWNIEEEGYQSVGSGSIFAKSSMKKLYSQVSDADSALKVAVEALYDAA
TH_0505|M.thermoresistible__bu AGGWNFEEEGYHAVGSGSIFAKSSMKKLYSQVTDADSALRVAVEALYDAA
MAB_2172|M.abscessus_ATCC_1997 AGGWHVESDGYQAVGSGSVFAKSSIKKLYRPGGDALTALAVAVEALYDAA
*** :.*.:**::*****:*****:**** *. :.: **:*******
Mb2134c|M.bovis_AF2122/97 DDDSATGGPDLVRGIFPTAVIIDADGAVDVPESRIAELARAIIESRSGAD
Rv2110c|M.tuberculosis_H37Rv DDDSATGGPDLVRGIFPTAVIIDADGAVDVPESRIAELARAIIESRSGAD
MMAR_3085|M.marinum_M DDDSATGGPDLVRGIYPTAVTINADGAVDVPEVRIAELAREVIGSRSRAD
MUL_2331|M.ulcerans_Agy99 DDDSATGGPDLVRGIYPTAVTINADGAVDVPEVRIAELAREVIGSRSRAD
MLBr_01322|M.leprae_Br4923 DDDSATGGPDLVRGIYPTAVTIGAEGAAEVTESRIAELAREIIESRSRAY
MAV_2404|M.avium_104 DDDSATGGPDLVRGIYPTAVTIGAEGALEVPESRIAELAREVIQSRSRAD
Mflv_3077|M.gilvum_PYR-GCK DDDSATGGPDLVRGIFPTAVVITADGAVEIAEQQIADMCREIIQNRSRAD
Mvan_3449|M.vanbaalenii_PYR-1 DDDSATGGPDLVRGIFPTAVVIGADGAHEVPEAKISSLCREVVENRSRTE
MSMEG_3895|M.smegmatis_MC2_155 DDDSATGGPDLVRGIYPTAVTIGAEGAEEVPETRIAELAREVIESRSRTD
TH_0505|M.thermoresistible__bu DDDSATGGPDLVRGIYPTAVIIDADGAEEVPESRIAELAREVIESRSRAD
MAB_2172|M.abscessus_ATCC_1997 DDDSATGGPDLVRRVFPTAIRIDSGGAVRVPEADIERIAREVIAKRTEAA
************* ::***: * : ** :.* * :.* :: .*: :
Mb2134c|M.bovis_AF2122/97 TFG--------SDGGEK
Rv2110c|M.tuberculosis_H37Rv TFG--------SDGGEK
MMAR_3085|M.marinum_M TFG--------PDGGEK
MUL_2331|M.ulcerans_Agy99 TFG--------PDGGEK
MLBr_01322|M.leprae_Br4923 TLG-------SFGGSEK
MAV_2404|M.avium_104 TFGP------DAAGGDK
Mflv_3077|M.gilvum_PYR-GCK TFGPDHAP---VRGDEL
Mvan_3449|M.vanbaalenii_PYR-1 TFGPDARP---TRGDEL
MSMEG_3895|M.smegmatis_MC2_155 TFGPDARRGIDARGDS-
TH_0505|M.thermoresistible__bu TFGPDAHRGIDARGDS-
MAB_2172|M.abscessus_ATCC_1997 REEADLRE---SGGDS-
*..