For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTQSSNVKTFDALFAELSERARTRPAGSGTVAALDGGVHGIGKKILEEAGEVWLAAEHESDDALAEEISQ LLYWTQVLMLARGLSLDDVYGKL
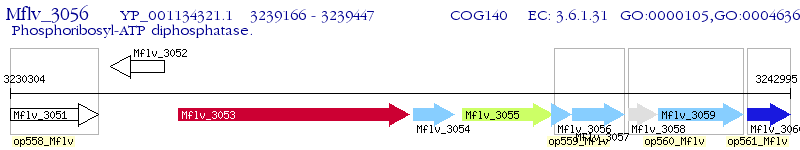
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3056 | hisE | - | 100% (93) | phosphoribosyl-ATP pyrophosphatase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2146c | hisE | 3e-39 | 82.80% (93) | phosphoribosyl-ATP pyrophosphatase |
| M. tuberculosis H37Rv | Rv2122c | hisE | 3e-39 | 82.80% (93) | phosphoribosyl-ATP pyrophosphatase |
| M. leprae Br4923 | MLBr_01309 | hisE | 3e-38 | 79.57% (93) | phosphoribosyl-ATP pyrophosphatase |
| M. abscessus ATCC 19977 | MAB_2131 | - | 7e-38 | 81.61% (87) | phosphoribosyl-ATP pyrophosphatase |
| M. marinum M | MMAR_3104 | hisE | 4e-37 | 79.12% (91) | phosphoribosyl-AMP pyrophosphatase HisE |
| M. avium 104 | MAV_2392 | hisE | 1e-41 | 88.17% (93) | phosphoribosyl-ATP pyrophosphatase |
| M. smegmatis MC2 155 | MSMEG_4181 | hisE | 9e-41 | 89.66% (87) | phosphoribosyl-ATP pyrophosphatase |
| M. thermoresistible (build 8) | TH_0038 | hisI | 2e-42 | 89.01% (91) | Probable phosphoribosyl-AMP pyrophosphatase HisE |
| M. ulcerans Agy99 | MUL_2353 | hisE | 3e-37 | 79.12% (91) | phosphoribosyl-ATP pyrophosphatase |
| M. vanbaalenii PYR-1 | Mvan_3471 | hisE | 6e-47 | 96.77% (93) | phosphoribosyl-ATP pyrophosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3104|M.marinum_M MKQSLAVKTFEDLFAELGDRARTRPPGSATVAALDAGVHTLGKKVLEEAG
MUL_2353|M.ulcerans_Agy99 MKQSLAVKTFEDLFAELGDRARTRPPGSATVAALDAGVHTLGKKVLEEAG
Mb2146c|M.bovis_AF2122/97 MQQSLAVKTFEDLFAELGDRARTRPADSTTVAALDGGVHALGKKLLEEAG
Rv2122c|M.tuberculosis_H37Rv MQQSLAVKTFEDLFAELGDRARTRPADSTTVAALDGGVHALGKKLLEEAG
MLBr_01309|M.leprae_Br4923 MKQSLAVKTFEDLFAELSERARTRPTDSATVASLDGGIHALGKKILEEAG
MAV_2392|M.avium_104 MKQSLAVKTFEDLFAELGERARTRPAGSATVAALDGGVHGLGKKILEEAG
Mflv_3056|M.gilvum_PYR-GCK MTQSSNVKTFDALFAELSERARTRPAGSGTVAALDGGVHGIGKKILEEAG
Mvan_3471|M.vanbaalenii_PYR-1 MTQSPNVKTFDALFAELSERARTRPAGSGTVAALDGGVHGIGKKILEEAG
MSMEG_4181|M.smegmatis_MC2_155 MKQFMLVKTFDALFAELSDKARTRPAGSGTVAALDAGVHGIGKKILEEAG
TH_0038|M.thermoresistible__bu MGQSIIVKTFDALFAELSDKARTRPAGSGTVAALDAGVHGIGKKVLEEAG
MAB_2131|M.abscessus_ATCC_1997 ------MKTFDELFAELSDRAKTRPEGSGTVAALDAGVHTLGKKLLEEAG
:***: *****.::*:*** .* ***:**.*:* :***:*****
MMAR_3104|M.marinum_M EVWLAAEHESNEALAEEISQLLYWTQVLMISRGVSLDEVYRKL
MUL_2353|M.ulcerans_Agy99 EVWLAAEHESNEALAEEISQLLYWTQVLMISRGVSLDEVYRKL
Mb2146c|M.bovis_AF2122/97 EVWLAAEHESNDALAEEISQLLYWTQVLMISRGLSLDDVYRKL
Rv2122c|M.tuberculosis_H37Rv EVWLAAEHESNDALAEEISQLLYWTQVLMISRGLSLDDVYRKL
MLBr_01309|M.leprae_Br4923 EVWLAAEHEPKEVLAEEISQLLYWTQVLMISRGLSLDDVYRKL
MAV_2392|M.avium_104 EVWLAAEHESDDALAGEISQLLYWTQVLMIARGLSLDDVYRKL
Mflv_3056|M.gilvum_PYR-GCK EVWLAAEHESDDALAEEISQLLYWTQVLMLARGLSLDDVYGKL
Mvan_3471|M.vanbaalenii_PYR-1 EVWLAAEHESDEALAEEISQLLYWTQVLMLSRGLSLDDVYGKL
MSMEG_4181|M.smegmatis_MC2_155 EVWLAAEHEGDEALAEEISQLLYWTQVLMVSRGLTLDDVYRKL
TH_0038|M.thermoresistible__bu EVWLAAEHESDDALAEEISQLLYWTQVLMIARGITLDDVYRKL
MAB_2131|M.abscessus_ATCC_1997 EVWLAAEHESDESLAEEVSQLLYWAQVLLIARGLTLDDVYRKL
********* .: ** *:******:***:::**::**:** **