For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARVRGFGELEASIMDRLWNRDPEADTTVRDIFDELSTERHIAYTTVMSTMDNLHGKGWLSRERDGKAYRY RPTLTREEHSARLMLEALSGGGRSEAVLSHFVAQIDAEESADLRAALRRLARKSGRR*
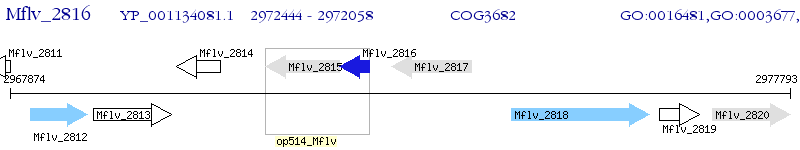
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2816 | - | - | 100% (128) | CopY family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0896 | - | 9e-33 | 57.76% (116) | CopY family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3357 | - | 8e-21 | 42.97% (128) | CopY family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1877c | - | 7e-20 | 40.15% (132) | transcriptional regulatory protein |
| M. tuberculosis H37Rv | Rv1846c | - | 5e-20 | 40.15% (132) | transcriptional regulatory protein |
| M. leprae Br4923 | MLBr_02063 | - | 3e-19 | 40.32% (124) | putative regulator |
| M. abscessus ATCC 19977 | MAB_4858 | - | 6e-41 | 69.64% (112) | penicillinase repressor |
| M. marinum M | MMAR_5060 | - | 8e-31 | 58.88% (107) | transcriptional regulatory protein |
| M. avium 104 | MAV_2869 | - | 2e-19 | 40.00% (125) | transcriptional repressor, CopY family protein |
| M. smegmatis MC2 155 | MSMEG_3630 | - | 5e-19 | 41.67% (120) | transcriptional repressor, CopY family protein |
| M. thermoresistible (build 8) | TH_1064 | - | 5e-18 | 40.50% (121) | POSSIBLE TRANSCRIPTIONAL REGULATORY PROTEIN |
| M. ulcerans Agy99 | MUL_4136 | - | 1e-34 | 58.82% (119) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_3706 | - | 2e-67 | 97.64% (127) | CopY family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1877c|M.bovis_AF2122/97 ------------------------------------------MAKLTRLG
Rv1846c|M.tuberculosis_H37Rv ------------------------------------------MAKLTRLG
MAV_2869|M.avium_104 ------------------------------------------MAKLTRLG
MLBr_02063|M.leprae_Br4923 ------------------------------------------MAKLTRLG
MSMEG_3630|M.smegmatis_MC2_155 ---------------------------------------------MTRLG
TH_1064|M.thermoresistible__bu ---------------------------------------------MTRLG
Mflv_2816|M.gilvum_PYR-GCK ------------------------------------------MARVRGFG
Mvan_3706|M.vanbaalenii_PYR-1 ---MAEAGWPLRDRSRRSASDSSTGTPVDTVSYLLTNSGGSIMARVRGFG
MAB_4858|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_5060|M.marinum_M --------------------------------------------------
MUL_4136|M.ulcerans_Agy99 MVLWSSVPGSPYLTEIKQARIGCISRLFGTGIAIDANVRESRAVQVRRFG
Mb1877c|M.bovis_AF2122/97 DLERAVMDHLWSRT--EPQTVRQVHEALSARRDLAYTTVMTVLQRLAKKN
Rv1846c|M.tuberculosis_H37Rv DLERAVMDHLWSRT--EPQTVRQVHEALSARRDLAYTTVMTVLQRLAKKN
MAV_2869|M.avium_104 DLERAVMDHLWSTA--EPQTVRQVHEALSAQRDLAYTTIMTVLQRLAKKN
MLBr_02063|M.leprae_Br4923 DLERAVMDHLWSRQ--EPQTVRQVHEALSARRDLAYTTVMTVLQRLAKKN
MSMEG_3630|M.smegmatis_MC2_155 ELEREVMDHLWSAP--EPQTVRQVHEALAARRDLAYTTIMTVLQRLAKKN
TH_1064|M.thermoresistible__bu DLEHEVMDHLWSAS--EPQTVRQVHEALAARRDLAYTTVMTVLHRLAKKK
Mflv_2816|M.gilvum_PYR-GCK ELEASIMDRLWNRDPEADTTVRDIFDELSTERHIAYTTVMSTMDNLHGKG
Mvan_3706|M.vanbaalenii_PYR-1 ELEASIMDRLWNRDPDADTTVRDIFDELNTERHIAYTTVMSTMDNLHGKG
MAB_4858|M.abscessus_ATCC_1997 ----MVMDRVWSRGDETATTVREVFDELAAERDIAYTTVMSTMDNLHTKG
MMAR_5060|M.marinum_M ------MDRVWDHG--GAVTVRGVFDDLAHDRRIAYTTVMSTMDNLHRKG
MUL_4136|M.ulcerans_Agy99 ELETVVMDRVWDHG--GAVTVRGVFDDLAHDRQIAYTTVMSTMDNLHRKG
**::*. *** :.: * * :****:*:.:..* *
Mb1877c|M.bovis_AF2122/97 LVLQIRDDRAHRYAPVHGRDELVAGLMVDALAQAEYSGSRQAALVHFVER
Rv1846c|M.tuberculosis_H37Rv LVLQIRDDRAHRYAPVHGRDELVAGLMVDALAQAEDSGSRQAALVHFVER
MAV_2869|M.avium_104 LVSQIRDDRAHRYAPVHGRDELVAGLMVDALAQAEDSGSRQAALVHFVER
MLBr_02063|M.leprae_Br4923 LVSQIRNNRAHRYAPVHGRDELVAGLMVDALDQAEDSDSRQAALVHFVER
MSMEG_3630|M.smegmatis_MC2_155 LVVQHRDDRAHRYAPTHGRDELVAGLMVDALDQAADSGSRQAALVHFVER
TH_1064|M.thermoresistible__bu LVVQHRDDRAHRYAPAYGRDELVAGLMVDALDQASDSGSRQAALVHFVER
Mflv_2816|M.gilvum_PYR-GCK WLSRERDGKAYRYRPTLTREEHSARLMLEALSGG---GRSEAVLSHFVAQ
Mvan_3706|M.vanbaalenii_PYR-1 WLSRERDGKAYRYRPTLTREEHSARLMLEALSGG---GRSEAVLSHFVAQ
MAB_4858|M.abscessus_ATCC_1997 WLEREREGKAYRYWAALTREQHSARLMREALSGG---GSPELVLTHFLEQ
MMAR_5060|M.marinum_M WLQRERVGKAFSYWPTMTREEHSARLMHDAFDAG---GDSDLVLAFFLKQ
MUL_4136|M.ulcerans_Agy99 WLQRERVGKAFSYWPTMTREEHSARLMHDAFDVG---GDSDLVLAFFLNQ
: : * .:*. * .. *:: * ** :*: . . : .* .*: :
Mb1877c|M.bovis_AF2122/97 VGADEADALRRALAELEAGHGNRPP-AGAATET---
Rv1846c|M.tuberculosis_H37Rv VGADEADALRRALAELEAGHGNRPP-AGAATET---
MAV_2869|M.avium_104 VGADEAEALRRALAELEANQRNNPASAGAGPEG---
MLBr_02063|M.leprae_Br4923 VGADEADALRRALAELETHQRHVRHLVALQRTTEGD
MSMEG_3630|M.smegmatis_MC2_155 VGVDEAAALRRALEELESKQRVLPPTGNSGTG----
TH_1064|M.thermoresistible__bu VGADEAEALRRALAELEAKHQDPPSTGGSGTD----
Mflv_2816|M.gilvum_PYR-GCK IDAEESADLRAALRRLARKSGRR-------------
Mvan_3706|M.vanbaalenii_PYR-1 IDAEESADLRAALRRLARKSRR--------------
MAB_4858|M.abscessus_ATCC_1997 ISAEESERLRAVLRRPAKRARTK-------------
MMAR_5060|M.marinum_M MDEHDAVRVRDALRRIIDDQEES-------------
MUL_4136|M.ulcerans_Agy99 MDEHDAVRVRDALRRIIDDQEES-------------
:. .:: :* .* .