For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAHRYKVREIAQQSGLSEATVDRVLNNRPGVRENTVAEVRQAIADLDKQRSQLRLNGRRYLIDVVMQTPQ RFSDAFRAAVEAELPAFAPAMVRARFHLWESGETAQMVEALGRLRGSHGVVLKAQDEPEVAEAIDRLVGS GVPVVTYTTDVPGSSRCGYVGIDNHGAGMTAAYLVQQWLGEATSGVLITLSRTAFRGEGEREVGFRSALR GSGRPVIEVSDSDGIDATNEALVLAALEANPTVEAVYSVGGGNVATVSAFERLNRVCRVFVAHDLDADNR RLLREGRISAVLHNDLRADARLAVRSILQERGALPVESTKPVPIQIVTPYNMPLHF
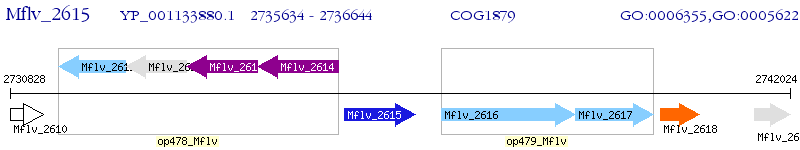
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2615 | - | - | 100% (336) | regulatory protein, LacI |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4648 | - | 1e-156 | 80.84% (334) | DNA-binding protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3966 | - | 1e-167 | 86.57% (335) | regulatory protein, LacI |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2615|M.gilvum_PYR-GCK MAHRYKVREIAQQSGLSEATVDRVLNNRPGVRENTVAEVRQAIADLDKQR
Mvan_3966|M.vanbaalenii_PYR-1 MAHRYKVREIAQQSGLSEATVDRVLHNRPGVRENTVAEVNQAIADLDKQR
MSMEG_4648|M.smegmatis_MC2_155 MAHRYKVREIAQQCGLSEATVDRVLNNRPGVRENTRAEVRQAIADLDKQR
*************.***********:********* ***.**********
Mflv_2615|M.gilvum_PYR-GCK SQLRLNGRRYLIDVVMQTPQRFSDAFRAAVEAELPAFAPAMVRARFHLWE
Mvan_3966|M.vanbaalenii_PYR-1 AQLRLNGRRYLIDVVMQTPRRFSDAFRAAVEAELPAFAPAMLRARFHLWE
MSMEG_4648|M.smegmatis_MC2_155 AQLRLNGRRFLIDVVMQTPRRFSDAFRTAVEAELPAFAPALLRARFHLWE
:********:*********:*******:************::********
Mflv_2615|M.gilvum_PYR-GCK SGETAQMVEALGRLRGSHGVVLKAQDEPEVAEAIDRLVGSGVPVVTYTTD
Mvan_3966|M.vanbaalenii_PYR-1 SGSTAQMVEALGRIRGSHGVVLKAQDDPAVAEAVDRLVDSGVPVVTYTTD
MSMEG_4648|M.smegmatis_MC2_155 SGSTAQMVDTLARIKSSHGVVLKAADEPEVVEAVDRLVAAGVPVVTYTTD
**.*****::*.*::.******** *:* *.**:**** :**********
Mflv_2615|M.gilvum_PYR-GCK VPGSSRCGYVGIDNHGAGMTAAYLVQQWLGEATSGVLITLSRTAFRGEGE
Mvan_3966|M.vanbaalenii_PYR-1 VPSSARCGYVGIDNHGAGVTAAYLVQQWLGEAPADVLITLSRTVFRGEGE
MSMEG_4648|M.smegmatis_MC2_155 VPTSARCAYVGIDNHGAGATAAYLIEQWLGADPADVLITLSRTVFRGEGE
** *:**.********** *****::**** .:.********.******
Mflv_2615|M.gilvum_PYR-GCK REVGFRSALRGSGRPVIEVSDSDGIDATNEALVLAALEANPTVEAVYSVG
Mvan_3966|M.vanbaalenii_PYR-1 REVGFRSALRNCGRTIVEVSDSDGIDATNERLVLDALAANPGVQAVYSVG
MSMEG_4648|M.smegmatis_MC2_155 REVGFRSGLRASGRTVVEVTDSDGIDATNERLVLQALERNPSVGAVYSPG
*******.** .**.::**:********** *** ** ** * **** *
Mflv_2615|M.gilvum_PYR-GCK GGNVATVSAFERLNRVCRVFVAHDLDADNRRLLREGRISAVLHNDLRADA
Mvan_3966|M.vanbaalenii_PYR-1 GGNVATVAAFEKIGRDCKVFIAHDLDADNRRLLRDGRISAVLHNDLRADA
MSMEG_4648|M.smegmatis_MC2_155 GGNTATVAAFDRLGRHCKVFIAHDLDVDNRRLLRDGRISAVLHNDLRADA
***.***:**:::.* *:**:*****.*******:***************
Mflv_2615|M.gilvum_PYR-GCK RLAVRSILQERGALPVESTKPVPIQIVTPYNMPLHF
Mvan_3966|M.vanbaalenii_PYR-1 RLALRLILQERGALPVEPVRPVPIQVVTPYNLPVHP
MSMEG_4648|M.smegmatis_MC2_155 RLAMRLILQQHGALPAEPVRPAPIQVVTPYNLPM--
***:* ***::****.*..:*.***:*****:*: