For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ALDDDDPVQRHLAVRPDSGYVYRTAWRVATGDIGSDLNLRLDGVARYIQEVGAENLVDAGEAEAHPHWLV QRTVIDVIEPIEFPNEVAFSRWCSGLSTRWCTMRVDLVGSDGGRIETEGFWIAMNAKTLTPQRATDSLIE RFSTTTTEHRLKWRPWLKNPADATESMPFALRRTDIDLFEHVTNTAYWHAIHEVIARVPDVCAPPYRAVI EYRKPIKFGEDVVITWRRDGDAPHVDIALTVDGEVRAAALLRTLSLG*
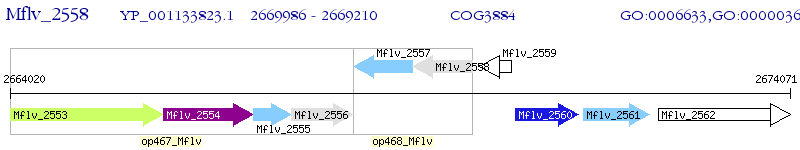
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2558 | - | - | 100% (258) | acyl-ACP thioesterase |
| M. gilvum PYR-GCK | Mflv_0108 | - | 8e-48 | 40.78% (206) | acyl-ACP thioesterase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2024 | - | 1e-84 | 62.30% (244) | hypothetical protein Mb2024 |
| M. tuberculosis H37Rv | Rv2001 | - | 4e-85 | 62.70% (244) | hypothetical protein Rv2001 |
| M. leprae Br4923 | MLBr_02463 | - | 4e-45 | 40.29% (206) | hypothetical protein MLBr_02463 |
| M. abscessus ATCC 19977 | MAB_4096c | - | 2e-45 | 39.61% (207) | hypothetical protein MAB_4096c |
| M. marinum M | MMAR_2977 | fatA | 3e-87 | 61.35% (251) | acyl-ACP thioesterase, FatA |
| M. avium 104 | MAV_4683 | - | 3e-47 | 42.51% (207) | acyl-ACP thioesterase superfamily protein |
| M. smegmatis MC2 155 | MSMEG_0909 | - | 2e-46 | 40.78% (206) | acyl-ACP thioesterase superfamily protein |
| M. thermoresistible (build 8) | TH_0977 | - | 1e-45 | 40.00% (210) | acyl-ACP thioesterase superfamily protein |
| M. ulcerans Agy99 | MUL_4535 | - | 4e-46 | 41.75% (206) | hypothetical protein MUL_4535 |
| M. vanbaalenii PYR-1 | Mvan_0800 | - | 3e-45 | 37.86% (206) | acyl-ACP thioesterase |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_02463|M.leprae_Br4923 ------------------MSLDKIMMPVPEGHPDVFDREWPLRVGDIDRT
MUL_4535|M.ulcerans_Agy99 ------------------MSLDKEMMPVPDGHPSVFDREWPLRVGDINRT
MAV_4683|M.avium_104 ------------------------MMPVPDGHPDVFDREWPLRVGDIDRT
TH_0977|M.thermoresistible__bu MAESTEKFAAGPVTGPARTGLGKALMPVPDPHPDVFDRQWPLRVADIDRT
MSMEG_0909|M.smegmatis_MC2_155 MTGTEKGSKDSSENSLVSTGLAKTMMPVPDPHPDVFDTGWPLRVADIDRN
Mvan_0800|M.vanbaalenii_PYR-1 MS-TTPG----------ATGLAKAMMPVPDPHPDVFDIQWPLRVADVDRE
MAB_4096c|M.abscessus_ATCC_199 ------------MSTEPSMGLDKDMMPVPHAHPHVYEARWPVRMADVDSG
Mb2024|M.bovis_AF2122/97 --------------MHHNRDVDLALVERPSSG-YVYTTGWRLATTDIDEH
Rv2001|M.tuberculosis_H37Rv --------------MHHNRDVDLALVERPSSG-YVYTTGWRLATTDIDEH
MMAR_2977|M.marinum_M --------------MPDNTDAQAPLVALPGAG-YVYRTSWRLATTDIDEH
Mflv_2558|M.gilvum_PYR-GCK ------------MALDDDDPVQRHLAVRPDSG-YVYRTAWRVATGDIGSD
: * *: * : *:.
MLBr_02463|M.leprae_Br4923 GRLRLDAGVRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPVEFQEM
MUL_4535|M.ulcerans_Agy99 GRLRLDAACRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPIEFQDM
MAV_4683|M.avium_104 GRLRLDAAARHIQDIGQDQLREMGFEETHPLWIVRRTMLDLIRPIEFNDM
TH_0977|M.thermoresistible__bu GRLRFDAACRHIQDIGQDQLRELGYEETHPLWIVRRTMIDLIRPIEFQDM
MSMEG_0909|M.smegmatis_MC2_155 GRLRFDASTRHIQDIGQDHLRQLGFEDTHPAWIVRRTMIDMIEPIEFPEL
Mvan_0800|M.vanbaalenii_PYR-1 GRLKFDAATRHIQDIGTDQLREMGYEDTHPLWIVRRTMIDMIEPVVFKDM
MAB_4096c|M.abscessus_ATCC_199 GRLRLDGAARHIQDIGQDHLRGVEAEETHPLWIVRRTMIDVIKPIDFKEA
Mb2024|M.bovis_AF2122/97 QQLRLDGVARYIQEVGAEHLADAQLAEVHPHWIVLRTVIDVINPIELPSD
Rv2001|M.tuberculosis_H37Rv QQLRLDGVARYIQEVGAEHLADAQLAEVHPHWIVLRTVIDVINPIELPSD
MMAR_2977|M.marinum_M MRLRLDGVARYIQEVGAEHLADAELAEVHPHWIVLRTVIDVIEPIELPSE
Mflv_2558|M.gilvum_PYR-GCK LNLRLDGVARYIQEVGAENLVDAGEAEAHPHWLVQRTVIDVIEPIEFPNE
.*::*. *:**::* ::* :.** *:* **::*:*.*: : .
MLBr_02463|M.leprae_Br4923 LRLRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWININRETQMPARIAD
MUL_4535|M.ulcerans_Agy99 LRCRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWIHVNKETEMPARIAD
MAV_4683|M.avium_104 LRLRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWININRETQMPSRIAD
TH_0977|M.thermoresistible__bu LRLRRWCSGTSNRWCEMRVRIDGRKGGLMESEAFWININRETQGPARISD
MSMEG_0909|M.smegmatis_MC2_155 LRLRRWCSGTSNRWCEMRVRIDGRKGGLVESEAFWININRETQGPSRIAD
Mvan_0800|M.vanbaalenii_PYR-1 LRLRRWCSGTSNRWCEMRVRIEGRKGGLIESEAFWININRETQGPARISD
MAB_4096c|M.abscessus_ATCC_199 LWLRRWCSATSNRWCQMRVRLDGRDGGLVESEAFWIHVSSETQGPARIDD
Mb2024|M.bovis_AF2122/97 ITFHRWCAALSTRWCSMRVQLQGSAGGHIETEGFWICVNKDTLTPSRLTD
Rv2001|M.tuberculosis_H37Rv ITFHRWCAALSTRWCSMRVQLQGSAGGRIETEGFWICVNKDTLTPSRLTD
MMAR_2977|M.marinum_M ITFRRWCAALSTRWCNMRVQLEGSHGGRIETEGFWLCVNKDTLTPSRLTD
Mflv_2558|M.gilvum_PYR-GCK VAFSRWCSGLSTRWCTMRVDLVGSDGGRIETEGFWIAMNAKTLTPQRATD
: ***:. *.*** *** : * ** :*:*.**: :. .* * * *
MLBr_02463|M.leprae_Br4923 DFLAGLHRTTSVDRLRWRGYLQPGSRDDASEIHEFPVRVTDIDLFDHMNN
MUL_4535|M.ulcerans_Agy99 DFLASLHKTTVVERLRWKGYLKPGSRDDATEIHEFPVRVTDIDLFDHMNN
MAV_4683|M.avium_104 DFLAGLHKTTSVDRLRWKPYLKPASRDDAVEIHEFPVRVTDIDLFDHMNN
TH_0977|M.thermoresistible__bu DFIAGLRRTTSVDRLRWQSYLRPGAREDAVQIQEFPVRASDIDLFDHMNN
MSMEG_0909|M.smegmatis_MC2_155 DFLAGLRRTTDIDRLRWKPYLKAGSREDALEIREYPVRVADIDLFDHMNN
Mvan_0800|M.vanbaalenii_PYR-1 DFIEGLRRTTDENRLRWKPYLRAGSREDAEHIRDYPVRVSDIDIFDHMNN
MAB_4096c|M.abscessus_ATCC_199 DFLATVASTTDVGRLRWKPYNKPGSRDTATDIRDFPVRFTDMDLFDHMNN
Mb2024|M.bovis_AF2122/97 DCIARFGSTTENHRLKWRPWLT--GPNIDGTETPFPLRRTDIDPFEHVNN
Rv2001|M.tuberculosis_H37Rv DCIARFGSTTENHRLKWRPWLT--GPNIDGTETPFPLRRTDIDPFEHVNN
MMAR_2977|M.marinum_M ACLTRFGSTTDDHRLKWRPWLT--EPITAGVDTPFPLRRTDIDPFEHVNN
Mflv_2558|M.gilvum_PYR-GCK SLIERFSTTTTEHRLKWRPWLK--NPADATESMPFALRRTDIDLFEHVTN
: . ** **:*: : :.:* :*:* *:*:.*
MLBr_02463|M.leprae_Br4923 SVYWSVIEDYLVSHSELLKGPLRTTIEHEAPVALGDKLEIVLHVHPAGST
MUL_4535|M.ulcerans_Agy99 SVYWSVIEDYLACHSELLQAPLRVTIEHEAPVALGDKLEIISHVHPAGST
MAV_4683|M.avium_104 SVYWSVVEDYLASHTELMRTPLRVTIEHEAPVALGDKLEIISHVHPAGST
TH_0977|M.thermoresistible__bu SVYWSVIEDYLFTQPELLRAPLRAAIEHNSPVALGDKLEIITHRHPPNST
MSMEG_0909|M.smegmatis_MC2_155 AVYWTVVEDYLYTHPELLAQPLRVTIEHDAPVALGDKLEIISHTHPPGTT
Mvan_0800|M.vanbaalenii_PYR-1 SVYWSVVEDYLYSQPELMSAPVRVTIEHDLPVALGDKLEIIRHVHPAGST
MAB_4096c|M.abscessus_ATCC_199 SVYWSIVEDHLSRESETRSAPFRVCLEHDSAVSLGDKLEIITNVYEDG--
Mb2024|M.bovis_AF2122/97 TIYWHGVHEILCQIPTLTAP-YRAVLEYRSPIKSGEPLTIRYEQHDDV--
Rv2001|M.tuberculosis_H37Rv TIYWHGVHEILCQIPTLTAP-YRAVLEYRSPIKSGEPLTIRYEQHDDV--
MMAR_2977|M.marinum_M TIYWHGIHEILSQQTTLTAP-YRAVLEYRSPIKFGETVTIRSEQRDGV--
Mflv_2558|M.gilvum_PYR-GCK TAYWHAIHEVIARVPDVCAPPYRAVIEYRKPIKFGEDVVITWRRDGDAP-
: ** :.: : . *. :*: .: *: : * .
MLBr_02463|M.leprae_Br4923 DQFGPGLVDRSVITLTYTVGDETKAIAAIFAL---
MUL_4535|M.ulcerans_Agy99 DKFGRGLTDRTVTTLTYAVGDETKAVAGLFSL---
MAV_4683|M.avium_104 DQFGPELADRTVTTLTYAVGDEAKAVAAIFAR---
TH_0977|M.thermoresistible__bu DQFGPELMDRTVTTLTYAVGDEVKAVAAVFAL---
MSMEG_0909|M.smegmatis_MC2_155 DKFGPELTDRTVTTLTYAVGEETKAVACLFSL---
Mvan_0800|M.vanbaalenii_PYR-1 DKFGEELSDRTVTTLTYAVGNETKAVAAIFPL---
MAB_4096c|M.abscessus_ATCC_199 TEFG--VPGRGVTTLTYVVGDEVKALASIFAR---
Mb2024|M.bovis_AF2122/97 ------------VRMHFVVGDDVRAAALLRRL---
Rv2001|M.tuberculosis_H37Rv ------------VRMHFVVGDDVRAAALLRRL---
MMAR_2977|M.marinum_M ------------THLQFTVGDDVRAAALLRKL---
Mflv_2558|M.gilvum_PYR-GCK -----------HVDIALTVDGEVRAAALLRTLSLG
: .*. :.:* * :