For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGTEKGSKDSSENSLVSTGLAKTMMPVPDPHPDVFDTGWPLRVADIDRNGRLRFDASTRHIQDIGQDHL RQLGFEDTHPAWIVRRTMIDMIEPIEFPELLRLRRWCSGTSNRWCEMRVRIDGRKGGLVESEAFWININR ETQGPSRIADDFLAGLRRTTDIDRLRWKPYLKAGSREDALEIREYPVRVADIDLFDHMNNAVYWTVVEDY LYTHPELLAQPLRVTIEHDAPVALGDKLEIISHTHPPGTTDKFGPELTDRTVTTLTYAVGEETKAVACLF SL
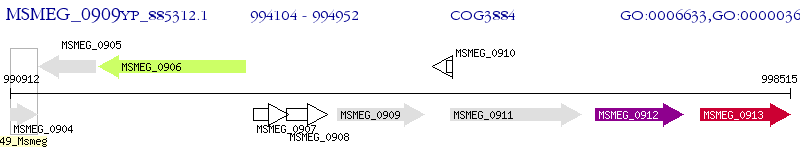
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0909 | - | - | 100% (282) | acyl-ACP thioesterase superfamily protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0475 | - | 1e-122 | 76.72% (262) | hypothetical protein Mb0475 |
| M. gilvum PYR-GCK | Mflv_0108 | - | 1e-126 | 77.57% (263) | acyl-ACP thioesterase |
| M. tuberculosis H37Rv | Rv0466 | - | 1e-122 | 76.72% (262) | hypothetical protein Rv0466 |
| M. leprae Br4923 | MLBr_02463 | - | 1e-123 | 75.95% (262) | hypothetical protein MLBr_02463 |
| M. abscessus ATCC 19977 | MAB_4096c | - | 5e-98 | 62.88% (264) | hypothetical protein MAB_4096c |
| M. marinum M | MMAR_0791 | - | 1e-122 | 77.10% (262) | hypothetical protein MMAR_0791 |
| M. avium 104 | MAV_4683 | - | 1e-126 | 80.16% (257) | acyl-ACP thioesterase superfamily protein |
| M. thermoresistible (build 8) | TH_0977 | - | 1e-129 | 78.41% (264) | acyl-ACP thioesterase superfamily protein |
| M. ulcerans Agy99 | MUL_4535 | - | 1e-122 | 77.10% (262) | hypothetical protein MUL_4535 |
| M. vanbaalenii PYR-1 | Mvan_0800 | - | 1e-132 | 80.38% (265) | acyl-ACP thioesterase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0108|M.gilvum_PYR-GCK MGTSQRVAGSGSVSTAGDGKQGLAKTLMPVPDPHPDVFDTQWPLRVADVD
Mvan_0800|M.vanbaalenii_PYR-1 MSTTP-------------GATGLAKAMMPVPDPHPDVFDIQWPLRVADVD
MSMEG_0909|M.smegmatis_MC2_155 MTGTEK--GSKDSSENSLVSTGLAKTMMPVPDPHPDVFDTGWPLRVADID
Mb0475|M.bovis_AF2122/97 --------------------MSLDKKLMPVPDGHPDVFDREWPLRVGDID
Rv0466|M.tuberculosis_H37Rv --------------------MSLDKKLMPVPDGHPDVFDREWPLRVGDID
MMAR_0791|M.marinum_M --------------------MSLDKEMMPVPDGHPSVFDREWPLRVGDIN
MUL_4535|M.ulcerans_Agy99 --------------------MSLDKEMMPVPDGHPSVFDREWPLRVGDIN
MLBr_02463|M.leprae_Br4923 --------------------MSLDKIMMPVPEGHPDVFDREWPLRVGDID
MAV_4683|M.avium_104 --------------------------MMPVPDGHPDVFDREWPLRVGDID
TH_0977|M.thermoresistible__bu MAESTE--KFAAGPVTGPARTGLGKALMPVPDPHPDVFDRQWPLRVADID
MAB_4096c|M.abscessus_ATCC_199 --------------MSTEPSMGLDKDMMPVPHAHPHVYEARWPVRMADVD
:****. ** *:: **:*:.*::
Mflv_0108|M.gilvum_PYR-GCK REGRLKFDAATRHIQDIGTDQLREMGYEETHPLWIVRRTMIDMVEPIVFK
Mvan_0800|M.vanbaalenii_PYR-1 REGRLKFDAATRHIQDIGTDQLREMGYEDTHPLWIVRRTMIDMIEPVVFK
MSMEG_0909|M.smegmatis_MC2_155 RNGRLRFDASTRHIQDIGQDHLRQLGFEDTHPAWIVRRTMIDMIEPIEFP
Mb0475|M.bovis_AF2122/97 RAGRLRLDAACRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPIEFG
Rv0466|M.tuberculosis_H37Rv RAGRLRLDAACRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPIEFG
MMAR_0791|M.marinum_M RTGRLRLDAACRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPIEFQ
MUL_4535|M.ulcerans_Agy99 RTGRLRLDAACRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPIEFQ
MLBr_02463|M.leprae_Br4923 RTGRLRLDAGVRHIQDIGQDQLREMGFEETHPLWIVRRTMVDLIRPVEFQ
MAV_4683|M.avium_104 RTGRLRLDAAARHIQDIGQDQLREMGFEETHPLWIVRRTMLDLIRPIEFN
TH_0977|M.thermoresistible__bu RTGRLRFDAACRHIQDIGQDQLRELGYEETHPLWIVRRTMIDLIRPIEFQ
MAB_4096c|M.abscessus_ATCC_199 SGGRLRLDGAARHIQDIGQDHLRGVEAEETHPLWIVRRTMIDVIKPIDFK
***::*.. ******* *:** : *:*** *******:*::.*: *
Mflv_0108|M.gilvum_PYR-GCK DILRLRRWCSGTSNRWCEMRVRIEGRRGGLVESEAFWININRETQGPARI
Mvan_0800|M.vanbaalenii_PYR-1 DMLRLRRWCSGTSNRWCEMRVRIEGRKGGLIESEAFWININRETQGPARI
MSMEG_0909|M.smegmatis_MC2_155 ELLRLRRWCSGTSNRWCEMRVRIDGRKGGLVESEAFWININRETQGPSRI
Mb0475|M.bovis_AF2122/97 DMLRCRRWCSGTSNRWCEMRVRVDGRKGGLIESEAFWIHVNRETEMPARI
Rv0466|M.tuberculosis_H37Rv DMLRCRRWCSGTSNRWCEMRVRVDGRKGGLIESEAFWIHVNRETEMPARI
MMAR_0791|M.marinum_M DMLRCRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWIHVNKETEMPARI
MUL_4535|M.ulcerans_Agy99 DMLRCRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWIHVNKETEMPARI
MLBr_02463|M.leprae_Br4923 EMLRLRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWININRETQMPARI
MAV_4683|M.avium_104 DMLRLRRWCSGTSNRWCEMRVRIDGRKGGLIESEAFWININRETQMPSRI
TH_0977|M.thermoresistible__bu DMLRLRRWCSGTSNRWCEMRVRIDGRKGGLMESEAFWININRETQGPARI
MAB_4096c|M.abscessus_ATCC_199 EALWLRRWCSATSNRWCQMRVRLDGRDGGLVESEAFWIHVSSETQGPARI
: * *****.******:****::** ***:*******::. **: *:**
Mflv_0108|M.gilvum_PYR-GCK SDDFIEGLRRTTDENRLRWKPYLKGGSREDAASVHRYPVRVSDIDIFDHM
Mvan_0800|M.vanbaalenii_PYR-1 SDDFIEGLRRTTDENRLRWKPYLRAGSREDAEHIRDYPVRVSDIDIFDHM
MSMEG_0909|M.smegmatis_MC2_155 ADDFLAGLRRTTDIDRLRWKPYLKAGSREDALEIREYPVRVADIDLFDHM
Mb0475|M.bovis_AF2122/97 ADDFLAGLHRTTSVDRLRWKGYLKPGSRDDASEIHEFPVRVTDIDLFDHM
Rv0466|M.tuberculosis_H37Rv ADDFLAGLHRTTSVDRLRWKGYLKPGSRDDASEIHEFPVRVTDIDLFDHM
MMAR_0791|M.marinum_M ADDFLASLHKTTAVERLRWKGYLKPGGRDDATEIHEFPVRVTDIDLFDHM
MUL_4535|M.ulcerans_Agy99 ADDFLASLHKTTVVERLRWKGYLKPGSRDDATEIHEFPVRVTDIDLFDHM
MLBr_02463|M.leprae_Br4923 ADDFLAGLHRTTSVDRLRWRGYLQPGSRDDASEIHEFPVRVTDIDLFDHM
MAV_4683|M.avium_104 ADDFLAGLHKTTSVDRLRWKPYLKPASRDDAVEIHEFPVRVTDIDLFDHM
TH_0977|M.thermoresistible__bu SDDFIAGLRRTTSVDRLRWQSYLRPGAREDAVQIQEFPVRASDIDLFDHM
MAB_4096c|M.abscessus_ATCC_199 DDDFLATVASTTDVGRLRWKPYNKPGSRDTATDIRDFPVRFTDMDLFDHM
***: : ** ****: * : ..*: * :: :*** :*:*:****
Mflv_0108|M.gilvum_PYR-GCK NNSVYWGVIEEYLNTRPDLMTAPLRVTIEHDLPVALGDDLEIIRHEYPAG
Mvan_0800|M.vanbaalenii_PYR-1 NNSVYWSVVEDYLYSQPELMSAPVRVTIEHDLPVALGDKLEIIRHVHPAG
MSMEG_0909|M.smegmatis_MC2_155 NNAVYWTVVEDYLYTHPELLAQPLRVTIEHDAPVALGDKLEIISHTHPPG
Mb0475|M.bovis_AF2122/97 NNAVYWSVIEDYLASHAELLRGPLRVTIEHEAPVALGDKLEIISHVHPAG
Rv0466|M.tuberculosis_H37Rv NNAVYWSVIEDYLASHAELLRGPLRVTIEHEAPVALGDKLEIISHVHPAG
MMAR_0791|M.marinum_M NNSVYWSVIEDYLACHSELLQAPLRVTIEHEAPVALGDKLEIISHVHPAG
MUL_4535|M.ulcerans_Agy99 NNSVYWSVIEDYLACHSELLQAPLRVTIEHEAPVALGDKLEIISHVHPAG
MLBr_02463|M.leprae_Br4923 NNSVYWSVIEDYLVSHSELLKGPLRTTIEHEAPVALGDKLEIVLHVHPAG
MAV_4683|M.avium_104 NNSVYWSVVEDYLASHTELMRTPLRVTIEHEAPVALGDKLEIISHVHPAG
TH_0977|M.thermoresistible__bu NNSVYWSVIEDYLFTQPELLRAPLRAAIEHNSPVALGDKLEIITHRHPPN
MAB_4096c|M.abscessus_ATCC_199 NNSVYWSIVEDHLSRESETRSAPFRVCLEHDSAVSLGDKLEIITNVYEDG
**:*** ::*::* ..: *.*. :**: .*:***.***: : : .
Mflv_0108|M.gilvum_PYR-GCK STDKFGAELADRGVTTLTYVVGDETKAVAAIFPL
Mvan_0800|M.vanbaalenii_PYR-1 STDKFGEELSDRTVTTLTYAVGNETKAVAAIFPL
MSMEG_0909|M.smegmatis_MC2_155 TTDKFGPELTDRTVTTLTYAVGEETKAVACLFSL
Mb0475|M.bovis_AF2122/97 STEIFGPGLVDRAVTTLTYVVGDEPKAVASLFNL
Rv0466|M.tuberculosis_H37Rv STEIFGPGLVDRAVTTLTYVVGDEPKAVASLFNL
MMAR_0791|M.marinum_M STDKFGPGLTDRTVTTLTYAVGDETKAVAGLFSL
MUL_4535|M.ulcerans_Agy99 STDKFGRGLTDRTVTTLTYAVGDETKAVAGLFSL
MLBr_02463|M.leprae_Br4923 STDQFGPGLVDRSVITLTYTVGDETKAIAAIFAL
MAV_4683|M.avium_104 STDQFGPELADRTVTTLTYAVGDEAKAVAAIFAR
TH_0977|M.thermoresistible__bu STDQFGPELMDRTVTTLTYAVGDEVKAVAAVFAL
MAB_4096c|M.abscessus_ATCC_199 --TEFG--VPGRGVTTLTYVVGDEVKALASIFAR
** : .* * ****.**:* **:* :*