For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TEALDTARLADWMDNSGLPGKGEPLSARFLSGGTQNVIFEIRRGEHRCVLRMPPPGAPPDRDKGILREWR IIEALDGTDVPHTAAVGVCADASVLGRPFYLMGYVDGWSPMDTHGTWPEPFNSDPSTRPGLSYQLAEGIA LLSKVDWRARGLHDLGRPDGFHERQVDRWIGFLDRIKNRELPGLDIATAWLRAHEPLDYVPGLMHGDYQF ANVMYRHGAPAQMAAIVDWEMGTVGDPKLDLGWMVQSWPEDTENPAPSEMGYVDMRGMPSRDDVVAHYAE VSGRQVDDLDYYLVLAKWKLAIVLEQGFQRAGDDEKLLAFGPVVTELMASAADLAESTDYRG*
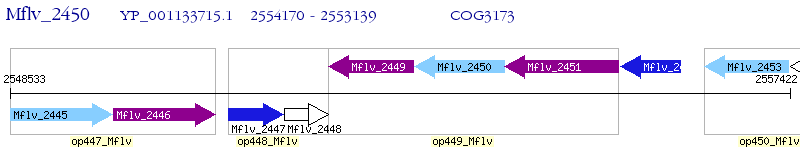
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2450 | - | - | 100% (343) | aminoglycoside phosphotransferase |
| M. gilvum PYR-GCK | Mflv_4318 | - | 1e-49 | 36.45% (310) | aminoglycoside phosphotransferase |
| M. gilvum PYR-GCK | Mflv_1204 | - | 3e-32 | 33.01% (306) | aminoglycoside phosphotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3787c | fadE36 | 2e-29 | 28.12% (313) | acyl-CoA dehydrogenase |
| M. tuberculosis H37Rv | Rv3761c | fadE36 | 2e-29 | 28.12% (313) | acyl-CoA dehydrogenase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4910c | - | 4e-60 | 39.76% (327) | putative aminoglycoside phosphotransferase |
| M. marinum M | MMAR_5309 | - | 3e-98 | 51.72% (348) | hypothetical protein MMAR_5309 |
| M. avium 104 | MAV_0312 | - | 1e-102 | 53.91% (345) | phosphotransferase enzyme family protein |
| M. smegmatis MC2 155 | MSMEG_4827 | - | 1e-173 | 83.92% (342) | aminoglycoside phosphotransferase |
| M. thermoresistible (build 8) | TH_4149 | - | 1e-170 | 81.10% (344) | PUTATIVE aminoglycoside phosphotransferase |
| M. ulcerans Agy99 | MUL_4383 | - | 4e-97 | 51.15% (348) | hypothetical protein MUL_4383 |
| M. vanbaalenii PYR-1 | Mvan_4201 | - | 0.0 | 91.25% (343) | aminoglycoside phosphotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2450|M.gilvum_PYR-GCK -------MTEALDTARLADWMDNSGLPGKGEPLSARFLSGGTQNVIFEIR
Mvan_4201|M.vanbaalenii_PYR-1 -------MSESLDTARLADWMDNSGHPGKGEPLAARFLSGGTQNVIFEIR
MSMEG_4827|M.smegmatis_MC2_155 ------MSTFQIDTMRLADWMDGAALPGKGEPLTARFLSGGTQNVIYELC
TH_4149|M.thermoresistible__bu ------VTEHRIDIVRLTAWMDEAALPGKGEPLEADFLSGGTQNVIYRIR
MMAR_5309|M.marinum_M METQGG---TSLNAAVLGRWLDSSHAPGSGEEPVLEQLEGGSQNTLYLVR
MUL_4383|M.ulcerans_Agy99 METEGG---TSLNAAVLGRWLDSRHAPGSGEEPVLEQLGGGSQNTLYLVR
MAV_0312|M.avium_104 MAGQRGGVESGLDAGVLARWLDANDAPGSGEETRLTPLQGGSQNTLYLIE
MAB_4910c|M.abscessus_ATCC_199 MAAPHHIDPKSVDFDVVSHWMDEQGLPG-GQLTDISAITGGTQNIMVRFS
Mb3787c|M.bovis_AF2122/97 --MTSVDRLDGLDLGALDRYLRSLGIGRDGELRG-ELISGGRSNLTFRVY
Rv3761c|M.tuberculosis_H37Rv --MTSVDRLDGLDLGALDRYLRSLGIGRDGELRG-ELISGGRSNLTFRVY
:: : :: *: : ** .* .
Mflv_2450|M.gilvum_PYR-GCK RGEHRCVLRMPPPGAPPDRDKGILREWRIIEALDGTDVPHTAAVGVCADA
Mvan_4201|M.vanbaalenii_PYR-1 RGDHHCVLRMPPPGAPSDRDKGILREWRIIEALDGTDVPHTAAVGVCTDA
MSMEG_4827|M.smegmatis_MC2_155 RGDARCVLRMPPPGAPSDRDKGILREWRIIAALDGTDVPHTEAVAMCDDT
TH_4149|M.thermoresistible__bu RGEHVCVLRMPPPDAPADRDKGILREWRIIEALDGTDVPHTAAVAVCEDS
MMAR_5309|M.marinum_M RGDQSMVLRMPGPRADAARIDGLLREIRLVRALSGTDVPHAELIAADETG
MUL_4383|M.ulcerans_Agy99 RGDQSMVLRMPGPRADAARIDGLLREIRLVRALSGTDVPHAELIATDETG
MAV_0312|M.avium_104 RGPARMVLRMPGARADAARIDGLLREIRLVRALSGTDVPHAALIAADDTG
MAB_4910c|M.abscessus_ATCC_199 RGGREYVLRRPPKHLREASNNVIRREARLLGALRGQGVPAPELIAACTDE
Mb3787c|M.bovis_AF2122/97 DDASSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTISLCQDD
Rv3761c|M.tuberculosis_H37Rv DDASSWLVRRPPLHGLTPSAHDMAREYRVVAALGDTPVPVARTISLCQDD
. ::* * . : ** *:: ** . ** . :.
Mflv_2450|M.gilvum_PYR-GCK SVLG-RPFYLMGYVDGWSPMDTHG-TWPEPFNSDPSTRPGLSYQLAEGIA
Mvan_4201|M.vanbaalenii_PYR-1 SVLG-RPFYLMGFVDGWSPMDTHG-KWPEPFDSDVSTRPGLSYQLAEGIA
MSMEG_4827|M.smegmatis_MC2_155 SVLG-RPFYLMGFVEGWSPMDRHG-VWPEPFDSDLSTRPGLSFQLAEGIA
TH_4149|M.thermoresistible__bu AVLG-RPFYLMGHVDGWSPMDLLDRRWPAPFDTDLQARAGLAYQLAEGIA
MMAR_5309|M.marinum_M SVLG-MPFYLMRAIDGWSPMDGG---WPAPFDTDLPARRGLALQLVEGAA
MUL_4383|M.ulcerans_Agy99 SVLG-MPFYLMRAIDGWSPMDGG---WPAPFDTDLPARRGLALQLVEGAA
MAV_0312|M.avium_104 TVLG-MPFYVMAAIDGWSPMDGG---WPAPFDTDPRARRGLAFQLVEGAA
MAB_4910c|M.abscessus_ATCC_199 TVLGGAVFYLMEPVDGFNASVG----LPELHAGNAEIRHQMGLEAVSGIA
Mb3787c|M.bovis_AF2122/97 SVLG-APFQVVEFVAGQVVRRRA----ELEALGSRSVIEGCVDALIRVLV
Rv3761c|M.tuberculosis_H37Rv SVLG-APFQVVEFVAGQVVRRRA----ELEALGSRSVIEGCVDALIRVLV
:*** * :: : * . .
Mflv_2450|M.gilvum_PYR-GCK LLSKVDWRARGLHDLGRPDGFHERQVDRWIGFLDRIKNRE------LPGL
Mvan_4201|M.vanbaalenii_PYR-1 LLSKVDWRAKGLQDLGRPDGFHERQVDRWIGFLERIKRRD------LPGL
MSMEG_4827|M.smegmatis_MC2_155 LLSKVDWRERGLADLGRPEGFHERQVDRWMAFLERIKRRE------LPGL
TH_4149|M.thermoresistible__bu LLAKVDWKARGLHDLGRPDGFHERQVDRWTGFFERIKGRE------LEGM
MMAR_5309|M.marinum_M KLGRVDWRGQGLEGFGRPDGFHERQVDRWLHFLAAYQVRE------LPGL
MUL_4383|M.ulcerans_Agy99 KLGRVDWRGQGLEGFGRPDGFHERQVDRWLHFLAAYQVRE------LPGL
MAV_0312|M.avium_104 KLGRVDWRKQGLEGFGRPDGFHERQVDRWLAFLDAYRVRE------LPGL
MAB_4910c|M.abscessus_ATCC_199 ALGALDYQELGLDGYGNPDGFLERQVPRWLKELDSYAKHDGYPGPDIPGL
Mb3787c|M.bovis_AF2122/97 DLHSIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHR---DADI
Rv3761c|M.tuberculosis_H37Rv DLHSIDPKAVGLSDFGKPDGYLERQVRRWGSQWELVRLPDDHR---DADI
* :* : ** . *.*:*: **** ** : .:
Mflv_2450|M.gilvum_PYR-GCK DIATAWLRAHEPLDYVPGLMHGDYQFANVMYRHGAPAQMAAIVDWEMGTV
Mvan_4201|M.vanbaalenii_PYR-1 EVATDWLRAHKPLDFIPGLMHGDYQFANVMYRHGAPAQLAAIVDWEMGTV
MSMEG_4827|M.smegmatis_MC2_155 AVATDWLRAHKPLDYIPGLMHGDYQFANVMYRHGAPATLAAIVDWEMGTV
TH_4149|M.thermoresistible__bu DVATEWLRTRRPRDYIPGLMHGDYQFANVMYHHGAPARLAAIVDWEMGTV
MMAR_5309|M.marinum_M DAASDWLRRNRPAHYTPGIMHGDYQFANVMFAHGEPARLAAIVDWEMTTI
MUL_4383|M.ulcerans_Agy99 DAASDWLRRNRPAHYTPGIMHGDYQFANVMFAHGEPARLAAIVDWAMTTI
MAV_0312|M.avium_104 DEASDWLRRNRPAHYRPGIMHGDYQFANVMFAHGEPARLAAIVDWEMTTV
MAB_4910c|M.abscessus_ATCC_199 QSVANWLERHRPSQWKPGIMHGDFHLANMMFRNDGP-QLAAIVDWEMSTI
Mb3787c|M.bovis_AF2122/97 SRLHLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTL
Rv3761c|M.tuberculosis_H37Rv SRLHLALQQAIPQQSRTSIVHGDYRIDNTILDTDDPCHVRAVVDWELSTL
*. * . ..::***::: * : . * : *:*** : *:
Mflv_2450|M.gilvum_PYR-GCK GDPKLDLGWMVQSWPEDTENPAPSEMGYVDMRGMPSRDDVVAHYAEVSGR
Mvan_4201|M.vanbaalenii_PYR-1 GDPKLDLGWMVQSWPEHTDAPEPSEMGYVDMRGMPSRDDVVAHYAEVSGR
MSMEG_4827|M.smegmatis_MC2_155 GDPKLDLAWMVQSWPTGADSGAGG-MDYVDMRGMPTRDDVVAHYAEVSGR
TH_4149|M.thermoresistible__bu GDPKLDLGWMVQSWPEDTADPQPSEMGYVDMRGMPSRDDVVAHYAEVSGR
MMAR_5309|M.marinum_M GDPLLDLAWSLLGY--DGEEPRAD-GFYLDMSAMPKRSELLEHYEKVSGL
MUL_4383|M.ulcerans_Agy99 GDSRLDLAWSLLGY--DGEEPRAD-GFYLDMSAMPKRSELLEHYEKVSGL
MAV_0312|M.avium_104 GDPLLDLAWCLLGY--DGENPRED-GFYLDMRGMPSRSELLEHYESVSGL
MAB_4910c|M.abscessus_ATCC_199 GDPLLDLGWMLATWPSETDDASLG-GALVQAGGLPTPQELVAHYAERTDR
Mb3787c|M.bovis_AF2122/97 GDPLSDAALMCVYRDPALDLIVHAQAAWTSP-LLPAADELADRYSLVSGQ
Rv3761c|M.tuberculosis_H37Rv GDPLSDAALMCVYRDPALDLIVHAQAAWTSP-LLPAADELADRYSLVSGQ
**. * . . :* .:: :* :.
Mflv_2450|M.gilvum_PYR-GCK QVDDLDYYLVLAKWKLAIVLEQGFQ---RAG------DDEKLLAFGPVVT
Mvan_4201|M.vanbaalenii_PYR-1 QVDDLDYYLVLAKWKLAIVLEQGFQ---RAG------DDEKLLAFGPVVT
MSMEG_4827|M.smegmatis_MC2_155 QVDDLDYYLVLAKWKLAIVLEQGFQ---RAG------DDEKLLAFGPVVT
TH_4149|M.thermoresistible__bu QVDDLDYYVILAKWKLAIVLEQGFQ---RAG------DNPRLQAYGPVIT
MMAR_5309|M.marinum_M STENIDYYLVLANWKLGIVLEKTYAAAMRAGPHAGGVDPKIKDAFAAMIP
MUL_4383|M.ulcerans_Agy99 STDNIDYYLVLANWKLGIVLEKTYAAAVRAGPHTGGVDPKIKDAFAAMIP
MAV_0312|M.avium_104 STENIDYYLVLANWKLGIVLEKTYAAGVRGGSRNEGVDPKITEAFGAMIP
MAB_4910c|M.abscessus_ATCC_199 DLGAMDWYTVLACFKLGIILEGTHAR-ACAG--------KAPVAVGDYLH
Mb3787c|M.bovis_AF2122/97 PLGHWEFYMALAYFKLAIIAAGIDYRRRMSEQAEG--KDTAAESVPDVVA
Rv3761c|M.tuberculosis_H37Rv PLGHWEFYMALAYFKLAIIAAGIDYRRRMSEQAEG--KDTAAESVPDVVA
::* ** :**.*: . : :
Mflv_2450|M.gilvum_PYR-GCK ELMASAADLAESTDYRG
Mvan_4201|M.vanbaalenii_PYR-1 GLMASAAELAESTDYRG
MSMEG_4827|M.smegmatis_MC2_155 GLMASAAELAESTDYRG
TH_4149|M.thermoresistible__bu ELMASAAELAESTDYR-
MMAR_5309|M.marinum_M RLIATAAELARATKAR-
MUL_4383|M.ulcerans_Agy99 RLIATAAELARATKAR-
MAV_0312|M.avium_104 QLIATAAELARQLGN--
MAB_4910c|M.abscessus_ATCC_199 GLTLQLFKRAVAITG--
Mb3787c|M.bovis_AF2122/97 PLIARGLAEIAKKSG--
Rv3761c|M.tuberculosis_H37Rv PLIARGLAEIAKKSG--
*