For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EPVELQTDRLMLRAVADADAAAIVDACNDPEILHYLPLPVPYTVGDARSFIAASAQEWVAGTRYGFGFFV MDTGELAGTCSLKPVTSGVLEVGYWSAAQHRRKGLTTEAVQRLCQWGFDVLDAHRIEWWAVVGNEGSRAV AEAAGFEMEGLLRQRACRRQEPADWWVGGLLPRPE*
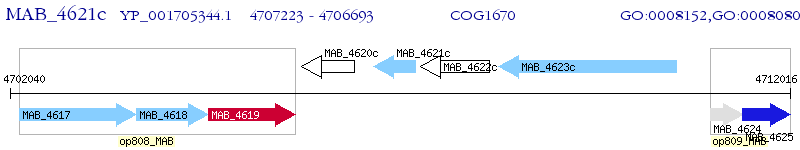
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_4621c | - | - | 100% (176) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_4838c | - | 2e-07 | 27.27% (132) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_1088 | - | 9e-06 | 30.95% (84) | ribosomal-protein-alanine acetyltransferase RimJ |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1022 | rimJ | 2e-05 | 33.33% (84) | ribosomal-protein-alanine acetyltransferase |
| M. gilvum PYR-GCK | Mflv_1899 | - | 2e-05 | 34.52% (84) | GCN5-related N-acetyltransferase |
| M. tuberculosis H37Rv | Rv0995 | rimJ | 2e-05 | 33.33% (84) | ribosomal-protein-alanine acetyltransferase |
| M. leprae Br4923 | MLBr_00184 | rimJ | 6e-06 | 33.33% (84) | putative acetyltransferase |
| M. marinum M | MMAR_4519 | rimJ | 7e-06 | 34.52% (84) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. avium 104 | MAV_3396 | - | 2e-05 | 30.12% (166) | acetyltransferase, gnat family protein |
| M. smegmatis MC2 155 | MSMEG_5469 | - | 6e-07 | 34.44% (90) | acetyltransferase, GNAT family protein |
| M. thermoresistible (build 8) | TH_0938 | rimJ | 8e-06 | 34.52% (84) | POSSIBLE RIBOSOMAL-PROTEIN-ALANINE ACETYLTRANSFERASE RIMJ |
| M. ulcerans Agy99 | MUL_4691 | rimJ | 5e-06 | 34.52% (84) | ribosomal-protein-alanine acetyltransferase RimJ |
| M. vanbaalenii PYR-1 | Mvan_4831 | - | 4e-06 | 35.71% (84) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4519|M.marinum_M ----MNLMRAGQRHPGWPMDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLA
MUL_4691|M.ulcerans_Agy99 ------------------MDVGPLRVRAGVIRLRPVRMRDGAQWSRTRLA
Mb1022|M.bovis_AF2122/97 ------------------MAVGPLRVSAGVIRLRPVRMRDGVHWSRIRLA
Rv0995|M.tuberculosis_H37Rv ------------------MAVGPLRVSAGVIRLRPVRMRDGVHWSRIRLA
MLBr_00184|M.leprae_Br4923 -------MRFNARHPGWPSNVGPLRVPAGVIRLRAVRLRDGVQWSRIRLA
Mflv_1899|M.gilvum_PYR-GCK ----MDLLSSRSQHPGWPAPVGPLRVSAGLVRLRPVRLRDAAQWSRIRLA
Mvan_4831|M.vanbaalenii_PYR-1 MAAVMNLLSSRSQHPGWPLAVGPLRVPAGVVRLRPVRLRDAAQWSRIRLA
MSMEG_5469|M.smegmatis_MC2_155 ------------MHPGWPDTVGPLRVPAGVVGLRPVRMRDAAAWSRIRLA
TH_0938|M.thermoresistible__bu ----VNLFRSSAAHPGWPIPAGPLRVKAGVIGLRPIRMRDAAVWSRLRLA
MAB_4621c|M.abscessus_ATCC_199 --------------------MEPVELQTDRLMLRAVADADAAAI--VDAC
MAV_3396|M.avium_104 ------------MNRTAMPEYPPDRITGPRLSLR-LPVLDDAGPLFQRVA
* .: : ** : * . .
MMAR_4519|M.marinum_M DRAHLEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDG
MUL_4691|M.ulcerans_Agy99 DRAHLEPWEPSAEGDWTIRHSVAAWPAVCSGLRSEARKGRMLPYVIELDG
Mb1022|M.bovis_AF2122/97 DRAHLEPWEPSADGEWTVRHTVAAWPAVCSGLRSEARNGRMLPYVIELDG
Rv0995|M.tuberculosis_H37Rv DRAHLEPWEPSADGEWTVRHTVAAWPAVCSGLRSEARNGRMLPYVIELDG
MLBr_00184|M.leprae_Br4923 DRAYLEPWEPSTEGDWVVRHSVIAWQVLCSSLRSEARKGRMLPYAIELDG
Mflv_1899|M.gilvum_PYR-GCK DRDHLEPWEPSTDMNWELRHSVSAWPSVCSGLRSEARKGRMLPYVIELDG
Mvan_4831|M.vanbaalenii_PYR-1 DRGHLEPWEPSTDMDWELRHSVSAWPSVCSGLRSEARKGRMLPYVIELDG
MSMEG_5469|M.smegmatis_MC2_155 DQHHLEPWEPMTGMDWKVRHAVTSWPSICSGLRAEARHGRMLPFVIELDG
TH_0938|M.thermoresistible__bu DRAHLEPWEPATGMDWETRHAISSWPSICSGLRAEARKGRLLPYVIELDG
MAB_4621c|M.abscessus_ATCC_199 NDPEILHYLPLP-VPYTVGDARSFIAASAQEWVAGTRYG--FGFFVMDTG
MAV_3396|M.avium_104 RDPQVTKY-----LLWAPHPDVAATRRVITEKLNASADEKTWVIELRHRG
: : : : : *
MMAR_4519|M.marinum_M RFCGQLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPV
MUL_4691|M.ulcerans_Agy99 RFCGQLTVGNVVHGALRSAWIGYWVASSQTGGGIATGAVALGLDHCFGPV
Mb1022|M.bovis_AF2122/97 QFCGQLTIGNVTHGALRSAWIGYWVPSAATGGGVATGALALGLDHCFGPV
Rv0995|M.tuberculosis_H37Rv QFCGQLTIGNVTHGALRSAWIGYWVPSAATGGGVATGALALGLDHCFGPV
MLBr_00184|M.leprae_Br4923 NFCGQLTIGNVTHGALRSAWIGYWVSSSATGGGVATGALALGLDHCFGPV
Mflv_1899|M.gilvum_PYR-GCK QFAGQLTIGNVTHGALRSAWIGYWVASGSTGGGVATAALALGLDHCFGPV
Mvan_4831|M.vanbaalenii_PYR-1 QFAGQLTIGNVTHGALRSAWIGYWVASGSTGGGVATAALALGLDHCFGPV
MSMEG_5469|M.smegmatis_MC2_155 EFVGQLTIGNVTHGALRSAWIGYWVASSRTGGGIATAALAMGLDHCFTAV
TH_0938|M.thermoresistible__bu RFVGQLTIGNVTHGALRSAWIGYWVASAVTGGGVATGALALGVDHCFSRV
MAB_4621c|M.abscessus_ATCC_199 ELAGTCSLKPVTSGVLE---VGYWSAAQHRRKGLTTEAVQRLCQWGFDVL
MAV_3396|M.avium_104 EVVGLLSCRRTAP---HSVEIGYCLGRRWWGKGLMSEVLGMLLTALDADR
.. * : .. . :** *: : .:
MMAR_4519|M.marinum_M GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLT
MUL_4691|M.ulcerans_Agy99 GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLVGLT
Mb1022|M.bovis_AF2122/97 MLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMAIT
Rv0995|M.tuberculosis_H37Rv MLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMAIT
MLBr_00184|M.leprae_Br4923 MLHRVEATVRPANVASRAVLAKVGFREEGLLRRYLEVDRAWRDHLLMALT
Mflv_1899|M.gilvum_PYR-GCK MLHRVEATVRPENGPSRAVLAKAGLREEGLLKRYLDVDGAWRDHLLVAIT
Mvan_4831|M.vanbaalenii_PYR-1 MLHRVEATVRPENGPSRAVLAKAGFREEGLLKRYLDVDGAWRDHLLVAIT
MSMEG_5469|M.smegmatis_MC2_155 QLHRIEATVRPENTPSRAVLAHVGFREEGLLKRYLEVDGAWRDHLLVAIT
TH_0938|M.thermoresistible__bu GLHRVEATVRPENAASRAVLAKVGFREEGLLRRYLDVDGAWRDHLLVAIT
MAB_4621c|M.abscessus_ATCC_199 DAHRIEWWAVVGNEGSRAVAEAAGFEMEGLLRQRACRRQEPADWWVGGLL
MAV_3396|M.avium_104 EVYRVWATCSVDNQRSARLLEHAGFFLEGRLARHAVYPTMGPEPQDSLLY
:*: * * : .*: ** * : : :
MMAR_4519|M.marinum_M VEEVEG-SVVSTLVRAGQASRL
MUL_4691|M.ulcerans_Agy99 VEEVEG-SVVSTLVRAGQASRL
Mb1022|M.bovis_AF2122/97 VEEVYG-SVASTLVRAGHASWP
Rv0995|M.tuberculosis_H37Rv VEEVYG-SVASTLVRAGHASWP
MLBr_00184|M.leprae_Br4923 IEEVSG-SVTSNLVRAGRASWL
Mflv_1899|M.gilvum_PYR-GCK AEEVNG-SVASALVRAGHASWA
Mvan_4831|M.vanbaalenii_PYR-1 IEELNG-TAASALVRAGRASWA
MSMEG_5469|M.smegmatis_MC2_155 AEELPQ-SAAHRLVAAGRAEWC
TH_0938|M.thermoresistible__bu VEELTSGSATAALVKHGRAEWV
MAB_4621c|M.abscessus_ATCC_199 PRPE------------------
MAV_3396|M.avium_104 AKILR-----------------
.