For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRFRAAAATALWLFAALGAAPSGFAQPVPAPDPAVPASAAAPPSPDAVASSPSGYLKTPDGWELTAGAKD ETQVAVAPLTTALTSREYVVGGTFTGTVTGAGSTKLSGGSLVVGYRIGCGIIGGPVEVLSSIGATTNFDQ SAVLGGVALPVNNQIKINLRPGTVTLVPVGEKKFKGSEARVTVTGFRIKTDGCVGQSFIQSYSTLTSSTT DTSDIVTYAGQVKVV
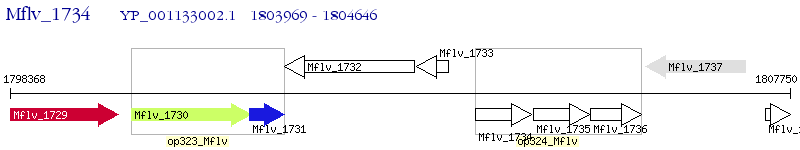
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1734 | - | - | 100% (225) | hypothetical protein Mflv_1734 |
| M. gilvum PYR-GCK | Mflv_1735 | - | 9e-56 | 53.30% (227) | hypothetical protein Mflv_1735 |
| M. gilvum PYR-GCK | Mflv_2295 | - | 1e-37 | 36.80% (250) | hypothetical protein Mflv_2295 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2799 | - | 4e-65 | 56.22% (217) | hypothetical protein MAB_2799 |
| M. marinum M | MMAR_1541 | - | 5e-39 | 39.32% (234) | hypothetical protein MMAR_1541 |
| M. avium 104 | MAV_3943 | - | 5e-42 | 41.56% (231) | hypothetical protein MAV_3943 |
| M. smegmatis MC2 155 | MSMEG_4360 | - | 7e-05 | 32.14% (140) | hypothetical protein MSMEG_4360 |
| M. thermoresistible (build 8) | TH_4651 | - | 3e-32 | 41.90% (179) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_2391 | - | 5e-39 | 39.32% (234) | hypothetical protein MUL_2391 |
| M. vanbaalenii PYR-1 | Mvan_5017 | - | 1e-90 | 71.81% (227) | hypothetical protein Mvan_5017 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1734|M.gilvum_PYR-GCK -----MRFRAAAATALWLFAALGAAPSGFAQP---VPAP--DPAVPASAA
Mvan_5017|M.vanbaalenii_PYR-1 -----MRIWSLVLAAICVLAPVGAMPRAVAQP---APEPGSPPAVDGVPA
MAB_2799|M.abscessus_ATCC_1997 -----MLKSLVSLTTVCALALT--APLAFADPEDPNPAPAPAPAQPVAND
MMAR_1541|M.marinum_M -MPISAVIRRAAVLAVCLVVSIS-APSAGAQP----------DAQPPP-G
MUL_2391|M.ulcerans_Agy99 MMPISAVIRRAAVLAVCLVVSIS-APSAGAQP----------DAQPPP-G
MAV_3943|M.avium_104 -----MVVRRALVAAVCLALSLPTAPTTGADP----------DAAPPPPE
TH_4651|M.thermoresistible__bu --------------------------------------------------
MSMEG_4360|M.smegmatis_MC2_155 ---MPAFRRLASTSRACVVSAVATTLVGGLVAG--LVGAPPAAADPCSVP
Mflv_1734|M.gilvum_PYR-GCK APPSPD-AVASSPSGYLKTPDGWELTAGAK---------DETQVAVAPLT
Mvan_5017|M.vanbaalenii_PYR-1 LPPSPA-AVPSSPPGYLATPDGWELTVAAR---------DETQVAVAPLT
MAB_2799|M.abscessus_ATCC_1997 VPPGPDGAIASAEPGNLITPDGWTVTVAGK---------NESLLPVAPLT
MMAR_1541|M.marinum_M AVPPDDGAVPPGTPAKITTPDGWVLALGAK---------DEKHVPVAPMT
MUL_2391|M.ulcerans_Agy99 AVPPDDGAVPPGTPAKITTPDGWVLALGAK---------DEKHVPVAPMT
MAV_3943|M.avium_104 AAPAAEGALPSNPPAILKTPDGWTLGLGAR---------DEQQVPVAPLT
TH_4651|M.thermoresistible__bu -----------------------VLTVWAK---------DETQIPIQPLT
MSMEG_4360|M.smegmatis_MC2_155 TPNASARGLTPSMPDIISKLPVLHLPIGRKPVQTASTPEEAKDTAVAPNT
: : : .: * *
Mflv_1734|M.gilvum_PYR-GCK TALTSREYVVG-GTFTGTVTGAG-----STKLSGGSLVVGYRIG------
Mvan_5017|M.vanbaalenii_PYR-1 TALTTREYLVG-GTFTGTVTGAG-----STELSGGSLVVGYRIG------
MAB_2799|M.abscessus_ATCC_1997 TALSSREYIVG-GSFIGAVSGAG-----KTKLAGGSLEAGYQIG------
MMAR_1541|M.marinum_M TAISSREYLSS-GIFGASLTGP--------ETPRGILEVGYEIG------
MUL_2391|M.ulcerans_Agy99 TAISSREYLSS-GIFGASLTGP--------ETPRGILEVGYEIG------
MAV_3943|M.avium_104 TALSSREYLAS-GIFVGSLSGP--------TQPQGVLEVGYEIG------
TH_4651|M.thermoresistible__bu TALSSREYEVG-ATFVGSITGPEG----SDETPEGVLEVGYEIG------
MSMEG_4360|M.smegmatis_MC2_155 AARAAAEPAASTATTVGWVTGPNSDSYSRFGISGADLGIMWDNGQTGANR
:* :: * . . . ::*. . . * : *
Mflv_1734|M.gilvum_PYR-GCK ------------CGIIGGP---VEVLSSIGATTN----FDQSAV------
Mvan_5017|M.vanbaalenii_PYR-1 ------------CGIVGGP---VELFATVGANPN----IGPNAV------
MAB_2799|M.abscessus_ATCC_1997 ------------CGIMGGP---VELTAGIGLSGG----FGTSG-------
MMAR_1541|M.marinum_M ------------CGIDMSTSNGFVMANSGGVAPS----IGAELPLGPGQP
MUL_2391|M.ulcerans_Agy99 ------------CGIDMSTSNGFVMANSGGVAPS----IGAELPLGPGQP
MAV_3943|M.avium_104 ------------CGIDMSTSDGVTIGGTAGITPG----VTGPVTGVPGD-
TH_4651|M.thermoresistible__bu ------------CGIDMSTSQGVSLSGSAGLNPSL-GLIATDIVAPDGLP
MSMEG_4360|M.smegmatis_MC2_155 QVLIAFGDTFGNCNVAAQEWRKNTLFRSADRNLADGMSIPNPVPGNIYAG
*.: : . .
Mflv_1734|M.gilvum_PYR-GCK ----LGGVALPVNNQIKINLRPGTVTLVP-----VGEKKFK--------G
Mvan_5017|M.vanbaalenii_PYR-1 ----LGAVEFPVNGQAKVNLRPGTVTVVP-----VGEKSFK--------G
MAB_2799|M.abscessus_ATCC_1997 ----LTSVSLPINGLVKVDLKPGQVTIVP-----VNKKSFE--------G
MMAR_1541|M.marinum_M PI-LIPLITGQYNNVVSVGLKPGFVVVVP-----VVRKEFK--------G
MUL_2391|M.ulcerans_Agy99 PI-LIPLITGQYNNVVSVGLKPGFVVVVP-----VVRKEFK--------G
MAV_3943|M.avium_104 ---VLPLVVAPIAGVLNVGLKPGLVIVVP-----VVKKQFR--------G
TH_4651|M.thermoresistible__bu IFGANAGIGPSLGGAVTVGLKPGFVNIVP-----VSKKPFK--------G
MSMEG_4360|M.smegmatis_MC2_155 SPVDAARPNFSRQVIKSLGLAGTEVTVIPTAGIAVGTTQYVNFMSVSRWG
.:.* * ::* * . : *
Mflv_1734|M.gilvum_PYR-GCK SEARVTVTGFRIKTDGCVGQSFIQSYSTLTSSTTDTSDIVTYAGQVKVV-
Mvan_5017|M.vanbaalenii_PYR-1 SDVRVTVTGFRVKVDGCVGQSFLQSYATLTGSTTDTSDIVTYAGQVKVV-
MAB_2799|M.abscessus_ATCC_1997 KTTRTTITGFRIKIDSCVGQSFIRSYATMTSSTDNTDDVVTYMGITKAV-
MMAR_1541|M.marinum_M ANPWIMISAFHLKIDGCVGQSFVRSYATLTRITDESDVVLSYVGSTKAV-
MUL_2391|M.ulcerans_Agy99 ANPWIMISAFYLKIDGCVGQSFVRSYATLTQITDESDVVLSYVGSTKAV-
MAV_3943|M.avium_104 PNPWVMVSNFHVKIDGCVGQSFIRSYAVLNRVTDESDVVLSYVGVTKAV-
TH_4651|M.thermoresistible__bu ADPWVMVKGFRVKIDGCVGESFIRSYAYLTRATDQSEAVLAWYGVTKKI-
MSMEG_4360|M.smegmatis_MC2_155 APGQWSTNFSAVAVSGDNGETWTVPATSIRPSWFNTVPGVPFVWGDQNFQ
. : .. *::: . : : :: :.: : .
Mflv_1734|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5017|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2799|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1541|M.marinum_M --------------------------------------------------
MUL_2391|M.ulcerans_Agy99 --------------------------------------------------
MAV_3943|M.avium_104 --------------------------------------------------
TH_4651|M.thermoresistible__bu --------------------------------------------------
MSMEG_4360|M.smegmatis_MC2_155 MAAYLRHNGYVYAYSTGAGRGGMPFLYRVVETAVADKSAYEYFTPFGWIK
Mflv_1734|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5017|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2799|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1541|M.marinum_M --------------------------------------------------
MUL_2391|M.ulcerans_Agy99 --------------------------------------------------
MAV_3943|M.avium_104 --------------------------------------------------
TH_4651|M.thermoresistible__bu --------------------------------------------------
MSMEG_4360|M.smegmatis_MC2_155 GFPFLALQVVWAPGSEMSVAWNDHLKKFVMLYTNTTSSVVMRTADKPEGP
Mflv_1734|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5017|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2799|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_1541|M.marinum_M --------------------------------------------------
MUL_2391|M.ulcerans_Agy99 --------------------------------------------------
MAV_3943|M.avium_104 --------------------------------------------------
TH_4651|M.thermoresistible__bu --------------------------------------------------
MSMEG_4360|M.smegmatis_MC2_155 WSRAKTIVSSTAVPGGIYAPYIHPWSEGKDLYFTLSRWSDYSVMLMRTSL
Mflv_1734|M.gilvum_PYR-GCK ---
Mvan_5017|M.vanbaalenii_PYR-1 ---
MAB_2799|M.abscessus_ATCC_1997 ---
MMAR_1541|M.marinum_M ---
MUL_2391|M.ulcerans_Agy99 ---
MAV_3943|M.avium_104 ---
TH_4651|M.thermoresistible__bu ---
MSMEG_4360|M.smegmatis_MC2_155 STS