For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
KRTRPTLALLAAVAVLTVGCGSRAGDEAAAPAAESCVDTTGDTVKVGAINSLSGGLAVSETVIRDAITLA VEQVNAGGGVLGKQLTLIGEDGASEPTIFAEKAQKLVQSDCVAAVFGGYTSASRKAMLPVFEDNNALLYY GQQYEGLESSPNIFYTGATTNQQIIPALDYLKEQGVKSLYLVGSDYVFPRTSNAIVKAYAQANGIEIKGE DYTPLGSTDFSTIVNKIRTADADAIFNVVVGDSLNAFFREYRSAGLTAEEMPVMSMCVGEEEVRSIGAPT LVGQLSSWNYYQTLDSPANTTFVEDFKARFGADRVTSDPMESAYAAVLLWKATVEKANSFAVPDIQEAAG GVTVDVPEGTMTLDGENHHVTKTARIGRVADDGLIYQVWQSPAPIEPDPYLKDYPWAAGITG*
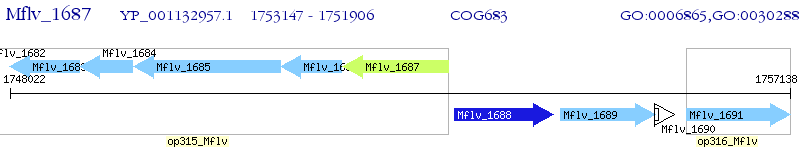
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1687 | - | - | 100% (413) | extracellular ligand-binding receptor |
| M. gilvum PYR-GCK | Mflv_3798 | - | e-169 | 69.23% (403) | extracellular ligand-binding receptor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2626c | - | 3e-06 | 20.58% (345) | ABC transporter ligand-binding protein |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_2982 | - | 1e-164 | 68.87% (408) | putative periplasmic binding protein |
| M. thermoresistible (build 8) | TH_2887 | - | 1e-173 | 72.46% (403) | PUTATIVE Extracellular ligand-binding receptor |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_2596 | - | 1e-172 | 70.72% (403) | extracellular ligand-binding receptor |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2982|M.smegmatis_MC2_155 MRVRGRATIRRSALATGSVITVAGLLLAGCGS------------------
Mvan_2596|M.vanbaalenii_PYR-1 -------------------MAAASLLLAGCGS------------------
TH_2887|M.thermoresistible__bu MRGPRRSRFNTSALRAGAVVFAAGLVLAGCGS------------------
Mflv_1687|M.gilvum_PYR-GCK ---------MKRTRPTLALLAAVAVLTVGCGS------------------
MAB_2626c|M.abscessus_ATCC_199 ----------MRARASVFMLGAVALALAGCHAGAQTDSSHQSNLKIVPLV
: ...: .** :
MSMEG_2982|M.smegmatis_MC2_155 ---KASETDAA-NAESCVDTSG------PTIKVGSLNSLSGTMAISEVTV
Mvan_2596|M.vanbaalenii_PYR-1 ---KASETDSA-NAESCVDTSG------STIKVGALNSLSGTMAISEVTV
TH_2887|M.thermoresistible__bu ---RAGDAPSS-GAASCVDTSG------PTIKVGSLNSLSGTMAISEVTV
Mflv_1687|M.gilvum_PYR-GCK ---RAGDEAAAPAAESCVDTTG------DTVKVGAINSLSGGLAVSETVI
MAB_2626c|M.abscessus_ATCC_199 QIAQDGSEVPASNTQSAADPAGDGKAQCANVSLATAGALNGPDSALGTNI
: .. .: : *..*.:* .:.:.: .:*.* : . :
MSMEG_2982|M.smegmatis_MC2_155 RDAIKLAVEEINADGGVLGKQIELIGEDGASEPTVFAEKAEKLISSDCVA
Mvan_2596|M.vanbaalenii_PYR-1 RNAIDLAVEQINASGGVMGKQIQLVGEDGASEPTVFAEKAEKLISSDCVA
TH_2887|M.thermoresistible__bu RDAIKLAVDEINADGGVLGKQIELIGEDGASEPTVFAEKAEKLISSDCVA
Mflv_1687|M.gilvum_PYR-GCK RDAITLAVEQVNAGGGVLGKQLTLIGEDGASEPTIFAEKAQKLVQSDCVA
MAB_2626c|M.abscessus_ATCC_199 RNGVQLAIDKHNAAN--PGCQIKLREFDTEGLPQNASNIAPQIIGDPTIV
*:.: **::: ** . * *: * * . * :: * ::: . :.
MSMEG_2982|M.smegmatis_MC2_155 AVFGGWTSSSRKAMLPVFESANALLYYPVQYEGLESSP--NIFYTGATTN
Mvan_2596|M.vanbaalenii_PYR-1 AVFGGWTSSSRKAMLPVFESANSLLYYPVQYEGLESSP--NIFYTGATTN
TH_2887|M.thermoresistible__bu AVFGGWTSSSRKAMLPVFESNNALLYYPVQYEGLESSP--NIFYTGATTN
Mflv_1687|M.gilvum_PYR-GCK AVFGGYTSASRKAMLPVFEDNNALLYYGQQYEGLESSP--NIFYTGATTN
MAB_2626c|M.abscessus_ATCC_199 GLIGPAFSGETKSTGQIFSEAGLVSTTGSATNVTLTQQGWKTFFRGLAND
.::* *.. *: :*.. . : : :. : *: * :.:
MSMEG_2982|M.smegmatis_MC2_155 QQIVPALD-YLKE-KGVKSLYLVGSDYVFPQ-TANRIIKAYAEANGIEIK
Mvan_2596|M.vanbaalenii_PYR-1 QQIVPALD-YLKE-KGIKSLYLVGSDYVFPQ-TANRIIKAYAAANGIEIK
TH_2887|M.thermoresistible__bu QQIVPALD-YLKE-QGVTSLYLVGSDYVFPQ-TANRIIKAYAPANGIEIK
Mflv_1687|M.gilvum_PYR-GCK QQIIPALD-YLKE-QGVKSLYLVGSDYVFPR-TSNAIVKAYAQANGIEIK
MAB_2626c|M.abscessus_ATCC_199 GVQGPAVANYLKKTLGYQKICVVDDSTDYGSGLANAVRGTLGNDCNVSVK
**: ***: * .: :*... : :* : : . .:.:*
MSMEG_2982|M.smegmatis_MC2_155 GEDYTPLGSTDFSTIVNKVRSANADAVFNTLNGDSNVAFFREYRNVGLTP
Mvan_2596|M.vanbaalenii_PYR-1 GEDYTPLGSTDFSTIVNKVRTADADAVFNTLNGDSNVAFFREYRNVGLTP
TH_2887|M.thermoresistible__bu GEDYTPLGSTDFSTIVNKIRTADADAVFNTLNGDSNVAFFREYRNVGLTA
Mflv_1687|M.gilvum_PYR-GCK GEDYTPLGSTDFSTIVNKIRTADADAIFNVVVGDSLNAFFREYRSAGLTA
MAB_2626c|M.abscessus_ATCC_199 K------GDKDFSAAVAQLKNAAPDAVYFGGYYAEAAPLVQQLRNGGVT-
*..***: * :::.* .**:: . .:.:: *. *:*
MSMEG_2982|M.smegmatis_MC2_155 DTMPVVSVSIAEEEVGGIGVQNIDGQLTAWNYYETIDTPENKAFVKAYKD
Mvan_2596|M.vanbaalenii_PYR-1 QDMPVVSVSIAEEEVGGIGVQNITGQLTAWNYYQTIDTPVNNEFVKAYKD
TH_2887|M.thermoresistible__bu QEMPVVSVSIAEEEIGGIGVQNIEGQLTAWNYYQTVDNPVNKAFVPAYKD
Mflv_1687|M.gilvum_PYR-GCK EEMPVMSMCVGEEEVRSIGAPTLVGQLSSWNYYQTLDSPANTTFVEDFKA
MAB_2626c|M.abscessus_ATCC_199 ------ATFVSSDGTKDVEFVKQAGDSAKDALLSCPCGPASNSFAKDYTA
: :..: .: . *: : . * .. *. :.
MSMEG_2982|M.smegmatis_MC2_155 FVKDPKKPTSDPMEAAYVSVYLWKNTVEKAQSFDVKAIQDNADGVSFDAP
Mvan_2596|M.vanbaalenii_PYR-1 KYG-ADKPTSDPMEAAYVSVYLWKNTVEKAQSFDVKAIQDNAGGVTFDAP
TH_2887|M.thermoresistible__bu EFG-AAKPTSDPMEAAYVSVYLWKNTVEKANSFDVDAIQENADGVSFDAP
Mflv_1687|M.gilvum_PYR-GCK RFG-ADRVTSDPMESAYAAVLLWKATVEKANSFAVPDIQEAAGGVTVDVP
MAB_2626c|M.abscessus_ATCC_199 KFN--QEPGTYSTEGYDLATILVKG-----------------------ID
. : . *. :. * *
MSMEG_2982|M.smegmatis_MC2_155 EGKVTIDGENHHITKTARIGEIRPDGLIYTVWESKGPIEPDPYLKSYPWA
Mvan_2596|M.vanbaalenii_PYR-1 EGLVTIDGENHHITKTARIGEIRPDGLIYTVWESPGPIEPDPYLKSYEWA
TH_2887|M.thermoresistible__bu EGTVTIDGETHHITKTARIGEIRPDGLIYTVWESPAPIEPDPYLKSYPWA
Mflv_1687|M.gilvum_PYR-GCK EGTMTLDGENHHVTKTARIGRVADDGLIYQVWQSPAPIEPDPYLKDYPWA
MAB_2626c|M.abscessus_ATCC_199 AGKSTRAGLLDWVRNYQGQGVAR-----EYKWNENGELTS-----TLIWV
* * * . : : * *:. . : . *.
MSMEG_2982|M.smegmatis_MC2_155 AGLSS
Mvan_2596|M.vanbaalenii_PYR-1 AGLSG
TH_2887|M.thermoresistible__bu AGLAS
Mflv_1687|M.gilvum_PYR-GCK AGITG
MAB_2626c|M.abscessus_ATCC_199 YKVT-
::