For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSVGGCRACAGTSEYQTVRLRFCGDSTFGGPRARTDARRGVRRAGRLRFCLRAAGGGRAGGSRRVQEAAL AQIGARRHRDADDRLRVRPVASRRGRGPGRFGARRLRTSRRRHRGYRRDRGDRRTSGSESGIGTGRSTRG RRPRPHHAHRRPGAGRSRTDDRRALHPAASARQVAGAGRGRVGGIADRRRCRPHRDHETRSAAASLTRRA APGAAGPPDRRRAAGAGLHRVGEPSAGPQRPRTDQRPDFRRVPRTRTTDRSAPHTAAAAGSPDADLHRPR HPPDADGRPVPQRVRGRLCARWDAGRAGSVHRQRVSAVTDQHLHRHGGQDLGDSRGPGDHRLPGHDGRIA SAVQPADHDHPGTRRSAGGVHPHDAARVLRRPRFRRAGARRGRGAGVRHRRHLRHRSLAGRMGGPVRRPA DRPGRHRFRGPRHEHPHAGQRRRLDVRQHRHVRELGAVHGDRSAGPATVHAALRARGRNETALRPDRDDR RPRGDDAAVQRIGAVGQGSDRHHDLPGGLPDQLLPVPPAGRAGLPPRCRPGPRHGAVPGHVAGPGPHRLR DVDHPAADEDGVRRRDPVPAVTFPGVTS
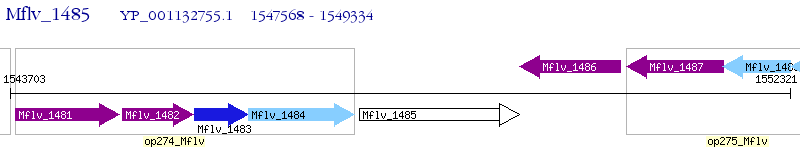
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1485 | - | - | 100% (588) | hypothetical protein Mflv_1485 |
| M. gilvum PYR-GCK | Mflv_2661 | - | 8e-05 | 31.25% (208) | hypothetical protein Mflv_2661 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_5011 | - | 2e-05 | 33.20% (247) | virulence factor Mce family protein |
| M. smegmatis MC2 155 | MSMEG_5152 | - | 8e-10 | 32.99% (194) | hypothetical protein MSMEG_5152 |
| M. thermoresistible (build 8) | TH_3755 | - | 8e-06 | 31.92% (213) | PUTATIVE cysteinyl-tRNA synthetase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1363 | - | 1e-08 | 30.23% (483) | LigA |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5152|M.smegmatis_MC2_155 ------------------------------MSNTSNSIRSSPDVQSIDWE
Mflv_1485|M.gilvum_PYR-GCK -----------MSVGGCRACAGTSEYQTVRLRFCGDSTFGGPRARTDARR
Mvan_1363|M.vanbaalenii_PYR-1 -------------MSRGSNHGNSSILVGRRRCGHRECCGGSPGDRTHHVH
TH_3755|M.thermoresistible__bu VTNSPAGLRLHDTMAGAVREFVPVRAGHVSIYLCGATVQGLPHIGHVRSG
MAV_5011|M.avium_104 -MGALRVIRRRSWQGLTLLVAAMVLTSCGWKGISNVSIPGGPGSGPNSYN
. *
MSMEG_5152|M.smegmatis_MC2_155 NDEIHLESHFHRHDRGSGRSPGRGGPERRGERRAGPG-------------
Mflv_1485|M.gilvum_PYR-GCK GVRRAGRLRFCLRAAGGGRAGGSRRVQEAALAQIGAR-------------
Mvan_1363|M.vanbaalenii_PYR-1 PGRGRRVCDGDPAQRGTGRQFAVVGVGTARHRRADGG-------------
TH_3755|M.thermoresistible__bu VAFDVLRRWLMAKGYDVAFIRNVTDIDDKILAKAAEAGRPWWEWAATYER
MAV_5011|M.avium_104 IYVQVPDTLAINGNSKVMVADVFVGSIKAIQLKNWIA-------------
:
MSMEG_5152|M.smegmatis_MC2_155 SARADAEPAGGQPTRRRAGRRAAAAAGRRLR-------------------
Mflv_1485|M.gilvum_PYR-GCK RHRDADDRLRVRPVASRRGRGPGRFGARRLRTSRRRHRGYRRDRGDRRTS
Mvan_1363|M.vanbaalenii_PYR-1 SAADQCGLRRVRRSRSGRVRRCADLRPNRTES------------------
TH_3755|M.thermoresistible__bu AFTAAYDALGVLPPSAEPRATGHITQMVELIERLLEKGHAYRANPDGDGD
MAV_5011|M.avium_104 TLTLGINKNVKLPKNATAKIGQTSLLGSQHVELAAPPNPSPELLKDGDTI
.
MSMEG_5152|M.smegmatis_MC2_155 ---------------------AHRNPGAGSRAAADR-----AGNPDPAA-
Mflv_1485|M.gilvum_PYR-GCK GSESGIGTGRSTRGRRPRPHHAHRRPGAGRSRTDDRRALHPAASARQVA-
Mvan_1363|M.vanbaalenii_PYR-1 ---------------------CCRRRIRVPRRTGGGVPRIGAGRGAGLS-
TH_3755|M.thermoresistible__bu VYFDVLSFPEYGRLSGHRIEDVHQGEGVATGKRDQRDFTLWKSAKPGEPS
MAV_5011|M.avium_104 PLKNSSSYPTTEQTLASLSLILRGGGIPNLEVLQNEVYNIFNGRGEAIRA
.
MSMEG_5152|M.smegmatis_MC2_155 -----GAGHS-------------------ARHGACRDRHRARGHPGRQRR
Mflv_1485|M.gilvum_PYR-GCK -----GAGR--------------------GRVGGIADRRRCRPHRDHETR
Mvan_1363|M.vanbaalenii_PYR-1 -----GRGG--------------------ERLGVRRRLRAVRSGRSHLRR
TH_3755|M.thermoresistible__bu WPTPWGRGRPGWHLECSAMAHTYLGSEFDIHCGGMDLVFPHHENEIAQSR
MAV_5011|M.avium_104 LLGKLDTFTN-------------------QLNQQRDDITHAIDSTNRLLT
.
MSMEG_5152|M.smegmatis_MC2_155 GL-------------------------------------ARAVGRGHHP-
Mflv_1485|M.gilvum_PYR-GCK SA-------------------------------------AASLTRRAAPG
Mvan_1363|M.vanbaalenii_PYR-1 RP-------------------------------------AGCGGVCLAAR
TH_3755|M.thermoresistible__bu AAGDGFAQYWLHNGWVTMGGEKMSKSLGNVLAIPAVLERVRAAELRYYLG
MAV_5011|M.avium_104 YVG-------------------------------GRADVVDRLLTDVPPL
.
MSMEG_5152|M.smegmatis_MC2_155 ------------AGARYADPAAADGQHPR---RPRFAGAHR---------
Mflv_1485|M.gilvum_PYR-GCK AAGPPDRRRAAGAGLHRVGEPSAGPQRPRTDQRPDFRRVPRT-------R
Mvan_1363|M.vanbaalenii_PYR-1 QR----------RGVRRGFRPVATDRRVLRRRPARCGGVR----------
TH_3755|M.thermoresistible__bu SAHYRSMLEFSETALQDAAKAYRGIEDFLHRVRIRVGGVEPGEWTPKFAA
MAV_5011|M.avium_104 IKHFADTKQLLINAVDSVGRLSQAADQYLSEARGPLHTDLQALQCP----
.
MSMEG_5152|M.smegmatis_MC2_155 ----------LAAGRPRPGRHQHRRQRGHGSRTRPHRTRG----------
Mflv_1485|M.gilvum_PYR-GCK TTDRSAPHTAAAAGSPDADLHRPRHPPDADGRPVPQRVRGRLCARWDAGR
Mvan_1363|M.vanbaalenii_PYR-1 -----------VAACQRRGFRRGLLPVGGDRRVLRRWTRC----------
TH_3755|M.thermoresistible__bu ALDDDLSVPIALAEVHHARAEGNRALNAGDHESALEHARSIRAMMGILGC
MAV_5011|M.avium_104 ------LKELGKASPYLIGALKLILTQPFDIDTVPKIFRG----------
* . . *
MSMEG_5152|M.smegmatis_MC2_155 ----------------------------FGPLGSRAAGAAADRRSAVTPE
Mflv_1485|M.gilvum_PYR-GCK AGSVHRQRVSAVTDQHLHRHGGQDLGDSRGPGDHRLPGHDGRIASAVQPA
Mvan_1363|M.vanbaalenii_PYR-1 -----------------------------GGRDRLTGAQRCGVRRELVPL
TH_3755|M.thermoresistible__bu DPLDERWETDRESSAALAALDVLVRWALEEREEARARRDWAQADTIRDRL
MAV_5011|M.avium_104 ---DYINISLTLDLTYSAVDNAFLTGTGLSGALRALEQSYGRDPETMIPD
MSMEG_5152|M.smegmatis_MC2_155 LV------------------------------------------------
Mflv_1485|M.gilvum_PYR-GCK DHDHPGTRRSAGGVHPHDAARVLRRPRFRRAGARRGRGAGVRHRRHLRHR
Mvan_1363|M.vanbaalenii_PYR-1 GCDRVVLRRLSRRRHHRHLRRHPVRVRRHHRNGRPGRRAGRRGGRGGGRR
TH_3755|M.thermoresistible__bu KEAGIEVTDTPDGPQWSLLDVTAVMXXXTSPYRAGPARRAAAAHLLPAAA
MAV_5011|M.avium_104 VRYTPNPNDAPGGPLVERGDRNCDSLRPTPTGRLRDPDRHLAARARHLLP
MSMEG_5152|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1485|M.gilvum_PYR-GCK SLAGRMGGPVRRPADRPGRHRFRGPRHEHPHAGQRRRLDVRQHRHVRELG
Mvan_1363|M.vanbaalenii_PYR-1 GPGGAGGGGAGRHGGSDGARSGGCGRHVRPRGGRRDRAG-RGFPGLRRRG
TH_3755|M.thermoresistible__bu ARSRRAPGGVGRADPQPARRAGRQRPRLLAGHPRHRRGAARRIRRQGRTG
MAV_5011|M.avium_104 ADPELGGDRPVHAQGRAAGVGRPVPDGQRHLSRHHHRQGHRRRAHRARRR
MSMEG_5152|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1485|M.gilvum_PYR-GCK AVHGDRSAGPA-TVHAALRARGRNETALRPDRDDRRPRGDDAAVQRIGAV
Mvan_1363|M.vanbaalenii_PYR-1 GRGAGGDHGAG-VERGHGTGGGFGAGAGRDHGGGGGFRDGRRGRDHGGGG
TH_3755|M.thermoresistible__bu AVAADRPADPAPARHPHRPAQSAPAAPGRRRGRRAVARRPDRGRLPGGHH
MAV_5011|M.avium_104 GDHEHRQPLQDPDRRGGQRALGVGGRRAVPGPGVERKPGQVPVVRADHHQ
MSMEG_5152|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1485|M.gilvum_PYR-GCK GQGSDRHHDLPGGLPDQLLPVPP-AGRAGLPPRCRPGPRHGAVPGHVAGP
Mvan_1363|M.vanbaalenii_PYR-1 GFRDGRRGRDHGGGGGFGVGVG----------RGRNLRHSGEVRHSGEVR
TH_3755|M.thermoresistible__bu LVRPGPRTVPAGAVPGRGRRRPAGAAATVELPPPRRPALDGARLRRRRHP
MAV_5011|M.avium_104 GHGAGRDRAGPGHREPRAGGAAQGEDRPVARRDGAVRRRVGPQPAAAGRR
MSMEG_5152|M.smegmatis_MC2_155 ----------------------------------
Mflv_1485|M.gilvum_PYR-GCK GPHRLRDVDHPAADEDGVRRRDPVPAVTFPGVTS
Mvan_1363|M.vanbaalenii_PYR-1 HGGEVGQCRHLGQGRSGVGERRPALTARP-----
TH_3755|M.thermoresistible__bu APLPAAGHRRPSDGVPGAGRRAAVTHRRPS----
MAV_5011|M.avium_104 HPGHRRRLQEPDHRRQRHHRALRAGDRQPGQIR-