For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TSANKIDGIMTNSTADASTTAAKRSDRVSNEAQRRRTFAVISHPDAGKSTLTEALVLHAKAITEAGAIHG KQGRRATVSDWMEMEKARGISITSTALQFPYTTQAGDTCVINLLDTPGHADFSEDTYRVLTAVDSAVMLI DAAKGLEPQTLKLFQVCAHRRIPIITVINKWDRPGRHALELMDEINERIGLRPTPLTWPVGIAGDFKGVL DRRTGKFIRFTRTAGGATAAPEEHIAAEAAHDAAGIDWDNAVEESELLEADGADFDPDSFLDCTSTPVLF TSAALNFGVNQLLDVLSALAPSPNGQLDVDGNRREAAAPFSAFVFKVQAGMDSAHRDRIAYARVCSGTFE RGDVLTHATTGKPFVTKYAQSVFGRERSTLDTAWPGDVIGLANANALRPGDTLYQDVAVQFPPIPSFSPE HFSVARGTDPSKHKQFRKGIEQLDQEGVVQVLRSDRRGDQAPVLAAVGPMQFEVAAHRMAAEFSAPIELE SLPYQVARICDPADAEALQKMAAVEVFTRTDGVMLAVFSTKWRLETVQRDNPDIMLRELIAAGQE*
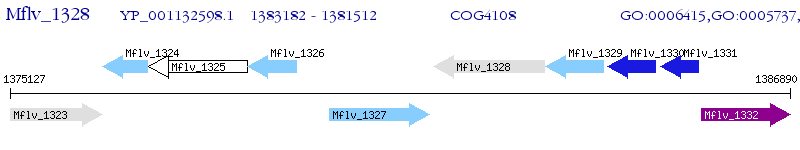
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1328 | - | - | 100% (556) | peptide chain release factor 3 |
| M. gilvum PYR-GCK | Mflv_5076 | - | 4e-37 | 26.63% (492) | elongation factor G |
| M. gilvum PYR-GCK | Mflv_2163 | - | 1e-27 | 28.64% (405) | GTP-binding protein TypA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0703 | fusA1 | 5e-36 | 27.52% (487) | elongation factor G |
| M. tuberculosis H37Rv | Rv0684 | fusA1 | 5e-36 | 27.52% (487) | elongation factor G |
| M. leprae Br4923 | MLBr_01878 | fusA | 3e-35 | 26.54% (486) | elongation factor G |
| M. abscessus ATCC 19977 | MAB_3849c | - | 3e-36 | 26.83% (492) | elongation factor G (EF-G) |
| M. marinum M | MMAR_1013 | fusA1 | 8e-36 | 27.07% (484) | elongation factor G, FusA1 |
| M. avium 104 | MAV_4490 | fusA | 6e-36 | 26.54% (486) | elongation factor G |
| M. smegmatis MC2 155 | MSMEG_1316 | prfC | 0.0 | 81.22% (543) | peptide chain release factor 3 |
| M. thermoresistible (build 8) | TH_1890 | fusA | 1e-36 | 27.76% (490) | PROBABLE ELONGATION FACTOR G FUSA1 (EF-G) |
| M. ulcerans Agy99 | MUL_0765 | fusA1 | 2e-36 | 27.27% (484) | elongation factor G |
| M. vanbaalenii PYR-1 | Mvan_5464 | - | 0.0 | 92.55% (537) | peptide chain release factor 3 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1013|M.marinum_M -----------------------MAQKDVLTDLTKVRNIGIMAHIDAGKT
MUL_0765|M.ulcerans_Agy99 -----------------------MAQKDVLTDLTKVRNIGIMAHIDAGKT
MAV_4490|M.avium_104 -----------------------MAQKDVLTDLTKVRNIGIMAHIDAGKT
MLBr_01878|M.leprae_Br4923 -----------------------MAQKDVLTDLTKVRNIGIMAHIDAGKT
Mb0703|M.bovis_AF2122/97 -----------------------MAQKDVLTDLSRVRNFGIMAHIDAGKT
Rv0684|M.tuberculosis_H37Rv -----------------------MAQKDVLTDLSRVRNFGIMAHIDAGKT
MAB_3849c|M.abscessus_ATCC_199 ------------------------MAQDVLTDLNKVRNIGIMAHIDAGKT
TH_1890|M.thermoresistible__bu -----------------------VAQKDVLTDLRRVRNIGIMAHIDAGKT
Mflv_1328|M.gilvum_PYR-GCK MTSANKIDGIMTNSTADASTTAAKRSDRVSNEAQRRRTFAVISHPDAGKS
Mvan_5464|M.vanbaalenii_PYR-1 ----------MT-----TSTAAGPRADLVSAEARRRRTFAVISHPDAGKS
MSMEG_1316|M.smegmatis_MC2_155 ----------MTDTPALAPSANTSLSRRIAAEATRRRTFAVISHPDAGKS
: : : *.:.:::* ****:
MMAR_1013|M.marinum_M TTTERILYYTGISYKIGEVHDG----AATMDWMEQEQERGITITSAATTC
MUL_0765|M.ulcerans_Agy99 TTTERILYYTGISYKIGEVHDG----AATMDWMEQEQERGITITSAATTC
MAV_4490|M.avium_104 TTTERILYYTGISYKIGEVHDG----AATMDWMEQEQERGITITSAATTC
MLBr_01878|M.leprae_Br4923 TTTERILYYTGISYKIGEVHDG----AATMDWMEQEQERGITITSAATTC
Mb0703|M.bovis_AF2122/97 TTTERILYYTGINYKIGEVHDG----AATMDWMEQEQERGITITSAATTT
Rv0684|M.tuberculosis_H37Rv TTTERILYYTGINYKIGEVHDG----AATMDWMEQEQERGITITSAATTT
MAB_3849c|M.abscessus_ATCC_199 TTTERILYYTGVNYKIGETHDG----ASTTDWMEQEQERGITITSAAVTC
TH_1890|M.thermoresistible__bu TTTERILFYTGVNYKMGETHDG----ASTTDWMEQEQERGITITSAAVTC
Mflv_1328|M.gilvum_PYR-GCK TLTEALVLHAKAITEAGAIHGKQGRRATVSDWMEMEKARGISITSTALQF
Mvan_5464|M.vanbaalenii_PYR-1 TLTEALVLHAKAITEAGAIHGKAGRRATVSDWMEMEKARGISITSTALQF
MSMEG_1316|M.smegmatis_MC2_155 TLTEALALHARVITEAGAIHGKAGRRSTVSDWMEMEKARGISITSTVLQF
* ** : :: : * *. ::. **** *: ***:***:.
MMAR_1013|M.marinum_M FWN-----DNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
MUL_0765|M.ulcerans_Agy99 FWN-----DNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
MAV_4490|M.avium_104 FWN-----DNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
MLBr_01878|M.leprae_Br4923 FWN-----DNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
Mb0703|M.bovis_AF2122/97 FWK-----DNQLNIIDTPGHVDFTVEVERNLRVLDGAVAVFDGKEGVEPQ
Rv0684|M.tuberculosis_H37Rv FWK-----DNQLNIIDTPGHVDFTVEVERNLRVLDGAVAVFDGKEGVEPQ
MAB_3849c|M.abscessus_ATCC_199 FWN-----GNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
TH_1890|M.thermoresistible__bu FWN-----DHQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQ
Mflv_1328|M.gilvum_PYR-GCK PYTTQAGDTCVINLLDTPGHADFSEDTYRVLTAVDSAVMLIDAAKGLEPQ
Mvan_5464|M.vanbaalenii_PYR-1 PYITQAGNTCVINLLDTPGHADFSEDTYRVLTAVDSAVMLIDAAKGLEPQ
MSMEG_1316|M.smegmatis_MC2_155 PYR-----DSVINLLDTPGHADFSEDTYRVLTAVDCAVMLIDAAKGLEPQ
: :*::*****.**: :. * * .:* ** ::*. :*:***
MMAR_1013|M.marinum_M SEQVWRQADKYDVPRICFVNKMDKIGADFYFSVRTMEERLGANVIPIQLP
MUL_0765|M.ulcerans_Agy99 SEQVWRQADKYDVPRICFVNKMDKIGADFYFSVRTMEERLGANVIPIQLP
MAV_4490|M.avium_104 SEQVWRQADKYDVPRICFVNKMDKIGADFYFSVRTMEERLGANVIPIQIP
MLBr_01878|M.leprae_Br4923 SEQVWRQADKYEVPRICFVNKMDKIGADFYFSVRTMQERLGANVIPIQLP
Mb0703|M.bovis_AF2122/97 SEQVWRQADKYDVPRICFVNKMDKIGADFYFSVRTMGERLGANAVPIQLP
Rv0684|M.tuberculosis_H37Rv SEQVWRQADKYDVPRICFVNKMDKIGADFYFSVRTMGERLGANAVPIQLP
MAB_3849c|M.abscessus_ATCC_199 SEQVWRQADKYDVPRICFVNKMDKLGADFYFTVQTIKDRLGAKPLVIQLP
TH_1890|M.thermoresistible__bu SEQVWRQADKYDVPRICFVNKMDKLGADFYFTVDTIKERLGANPLVIQLP
Mflv_1328|M.gilvum_PYR-GCK TLKLFQVCAHRRIPIITVINKWDRPGRHALELMDEINERIGLRPTPLTWP
Mvan_5464|M.vanbaalenii_PYR-1 TLKLFQVCRHRRIPIITVVNKWDRPGRHALELMDEIQSQIGLRPTPLTWP
MSMEG_1316|M.smegmatis_MC2_155 TLKLFQVCKHRGIPIITVINKWDRPGRHALELIDEIQTRIGLKPTPLTWP
: :::: . : :* * .:** *: * . : : ::* . : *
MMAR_1013|M.marinum_M VGSEGDFEGVVDLVEMKAKVWSADAKLGEKYDVVDIPADLQEKADEYRTK
MUL_0765|M.ulcerans_Agy99 VGSEGDFEGVVDLVEMKAKVWSADAKLGEKYDVVDIPADLQEKADEYRTK
MAV_4490|M.avium_104 VGSEGDFEGVVDLVEMKAKVWSAEAKLGEKYDVVDIPADLQEKAEEYRTK
MLBr_01878|M.leprae_Br4923 VGSEGDFEGVVDLVEMKAKVWSTEAKLGEKYDVVGIPTDLQEKAEEYRTN
Mb0703|M.bovis_AF2122/97 VGAEADFEGVVDLVEMNAKVWRGETKLGETYDTVEIPADLAEQAEEYRTK
Rv0684|M.tuberculosis_H37Rv VGAEADFEGVVDLVEMNAKVWRGETKLGETYDTVEIPADLAEQAEEYRTK
MAB_3849c|M.abscessus_ATCC_199 IGAENDFEGIIDLVEMNAKVWRGETKLGESYETVEIPADLADKAAEYRNE
TH_1890|M.thermoresistible__bu IGAENDFVGVVDLVEMKAKVWRGETALGEKYEVEEIPAELADKAEEYRNA
Mflv_1328|M.gilvum_PYR-GCK VGIAGDFKGVLDRRTGKFIRFTRTAGGATAAPEEHIAAEAAHDAAG--ID
Mvan_5464|M.vanbaalenii_PYR-1 VGIAGDFKGVLDRRTGKFIRFTRTAGGATAAPEEHIDPDKAHDAAG--VD
MSMEG_1316|M.smegmatis_MC2_155 VGIAGDFKGVLDRRGGQFIRFTRTAGGATAAPEEHIPADAAHAAAG--DD
:* ** *::* : : : . * .: . *
MMAR_1013|M.marinum_M LLEAVAETDEALLEKYLGGEELTEAEIKGAIRKLTITSEAYPVLCGSAFK
MUL_0765|M.ulcerans_Agy99 LLEAVAETDEALLEKYLGGEELTEAEIKGAIRKLTITSEAYPVLCGSAFK
MAV_4490|M.avium_104 LLEAVAETDEALLDKYLGGEELTIEEIKGAIRKLTISSEAYPVLCGSAFK
MLBr_01878|M.leprae_Br4923 LLETVAETDEALLEKYFSGEELTVAEIKGAIRKLTISSEAYPVLCGSAFK
Mb0703|M.bovis_AF2122/97 LLEVVAESDEHLLEKYLGGEELTVDEIKGAIRKLTIASEIYPVLCGSAFK
Rv0684|M.tuberculosis_H37Rv LLEVVAESDEHLLEKYLGGEELTVDEIKGAIRKLTIASEIYPVLCGSAFK
MAB_3849c|M.abscessus_ATCC_199 LLETVAESDEALLEKYLGGEELSIDEIKAGIRKLTVASELYPVLCGSAFK
TH_1890|M.thermoresistible__bu LLEAVAETDEDLLEKYLGGEELTVEEIKGAIRKLTVTSQAYPVLCGSAFK
Mflv_1328|M.gilvum_PYR-GCK WDNAVEES------------ELLEADGADFDPDSFLDCTSTPVLFTSAAL
Mvan_5464|M.vanbaalenii_PYR-1 WDNAVEES------------ELLSADGADFDPDTFLDCTSTPVLFTSAAL
MSMEG_1316|M.smegmatis_MC2_155 WDTAVEES------------ELLSADGGDYDRDAFLAGTASPTLFTSAAL
.* *: ** : . : *.* **
MMAR_1013|M.marinum_M NKGVQPMLDAVIDYLPSPLDVPAAIGHVPGKEDEEVVRKPSTDEPFSALA
MUL_0765|M.ulcerans_Agy99 NKGVQPMLDAVIDYLPSPLDVPAAIGHVPGKEDEEVVRKPSTDEPFSALA
MAV_4490|M.avium_104 NKGVQPMLDAVIDYLPSPLDVPPAEGHVPGKEEELITRKPSTDEPFSALA
MLBr_01878|M.leprae_Br4923 NKGVQPMLDAVIDYLPSPLDVPAAIGHVPGKEDEEIVRKPSTDEPLSALA
Mb0703|M.bovis_AF2122/97 NKGVQPMLDAVVDYLPSPLDVPPAIGHAPAKEDEEVVRKATTDEPFAALA
Rv0684|M.tuberculosis_H37Rv NKGVQPMLDAVVDYLPSPLDVPPAIGHAPAKEDEEVVRKATTDEPFAALA
MAB_3849c|M.abscessus_ATCC_199 NKGVQPMLDAVIDYLPSPLDVESVKGHVPGHEDQEIERKPSTDEPFSALA
TH_1890|M.thermoresistible__bu NKGVQPMLDAVIDYLPSPLDVESVKGHVPGHEDQEIARKPSVDEPFSALA
Mflv_1328|M.gilvum_PYR-GCK NFGVNQLLDVLSALAPSPNGQLDVDG-----------NRREAAAPFSAFV
Mvan_5464|M.vanbaalenii_PYR-1 NFGVNQLLDVLAQLAPSPNGALDVDG-----------NRREAAAPFSAFV
MSMEG_1316|M.smegmatis_MC2_155 NFGVNQLLDVLVELAPSPNGALDVDG-----------ERRDPDAPFSAFV
* **: :**.: *** . . * .: *::*:.
MMAR_1013|M.marinum_M FKVAT---HPFFGKLTYVRVYSGKVDSGSQVINSTKGKKERLGKLFQMHS
MUL_0765|M.ulcerans_Agy99 FKVAT---HPFFGKLTYVRVYSGKVDSGSQVINSTKGKKERLGKLFQMHS
MAV_4490|M.avium_104 FKVAT---HPFFGKLTYVRVYSGKVDSGSQVINSTKGKKERLGKLFQMHS
MLBr_01878|M.leprae_Br4923 FKVAT---HPFFGKLTYVRVYSGKVDSGSQVINATKGKKERLGKLFQMHS
Mb0703|M.bovis_AF2122/97 FKIAT---HPFFGKLTYIRVYSGTVESGSQVINATKGKKERLGKLFQMHS
Rv0684|M.tuberculosis_H37Rv FKIAT---HPFFGKLTYIRVYSGTVESGSQVINATKGKKERLGKLFQMHS
MAB_3849c|M.abscessus_ATCC_199 FKIAV---HPFFGKLTYVRVYSGKIESGAQVVNATKGKKERLGKLFQMHA
TH_1890|M.thermoresistible__bu FKIAV---HPFFGKLTYIRVYSGQVESGSQVMNSTKGKKERLGKLFQMHS
Mflv_1328|M.gilvum_PYR-GCK FKVQAGMDSAHRDRIAYARVCSGTFERGDVLTHATTGKPFVTKYAQSVFG
Mvan_5464|M.vanbaalenii_PYR-1 FKVQAGMDSAHRDRIAYARVCSGTFERGDVLTHASTGKPFVTKYAQSVFG
MSMEG_1316|M.smegmatis_MC2_155 FKVQAGMDSAHRDRIAYARVVSGTFERGDVLTHAATGKPFVTKYAQSVFG
**: . .. .:::* ** ** .: * : :::.** .:..
MMAR_1013|M.marinum_M NKENPVETASAGHIYAVIGLKDTTTGDTLSDPNNQIVLESMTFPDPVIEV
MUL_0765|M.ulcerans_Agy99 NKESPVETASAGHIYAVIGLKDTTTGDTLSDPNNQIVLESMTFPDPVIEV
MAV_4490|M.avium_104 NKENPVETASAGHIYAVIGLKDTTTGDTLSDPNHQIVLESMTFPDPVIEV
MLBr_01878|M.leprae_Br4923 NKENPVETASAGHIYAVIGLKDTTTGDTLADPNNQIVLESMTFPDPVIEV
Mb0703|M.bovis_AF2122/97 NKENPVDRASAGHIYAVIGLKDTTTGDTLSDPNQQIVLESMTFPDPVIEV
Rv0684|M.tuberculosis_H37Rv NKENPVDRASAGHIYAVIGLKDTTTGDTLSDPNQQIVLESMTFPDPVIEV
MAB_3849c|M.abscessus_ATCC_199 NKENPVETAAAGHIYAVIGLKDTTTGDTLCDPNSQIVLESMTFPDPVIEV
TH_1890|M.thermoresistible__bu NKENPVERVSAGHIYAVIGLKDTTTGDTLCDPNDQIVLESMTFPDPVIEV
Mflv_1328|M.gilvum_PYR-GCK RERSTLDTAWPGDVIGLANANALRPGDTLYQDVAVQFPPIPSFSPEHFSV
Mvan_5464|M.vanbaalenii_PYR-1 RERSTLDTAWPGDVIGLANAAALRPGDTLYQDIAVQYPPIPSFSPEHFSV
MSMEG_1316|M.smegmatis_MC2_155 QQRSTLDDAWPGDVIGLANAQALRPGDTLYRDVPVQFPPIPSFSPEHFAV
.:...:: . .*.: .: . .**** :*. : *
MMAR_1013|M.marinum_M AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQT-VIGGMGELHLD
MUL_0765|M.ulcerans_Agy99 AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQT-VIGGMGELHLD
MAV_4490|M.avium_104 AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQT-VIGGMGELHLD
MLBr_01878|M.leprae_Br4923 AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQT-VIGGMGELHLD
Mb0703|M.bovis_AF2122/97 AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQT-VIGGMGELHLD
Rv0684|M.tuberculosis_H37Rv AIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQT-VIGGMGELHLD
MAB_3849c|M.abscessus_ATCC_199 AIEPKTKTDQEKLGTAIQKLAEEDPTFKVKLDQETGQT-VIGGMGELHLD
TH_1890|M.thermoresistible__bu AIEPKTKSDQEKLGTAIQKLAEEDPTFKVHQDPETGQT-VIGGMGELHLD
Mflv_1328|M.gilvum_PYR-GCK ARGTDP-SKHKQFRKGIEQLDQEGVVQVLRSDRRGDQAPVLAAVGPMQFE
Mvan_5464|M.vanbaalenii_PYR-1 ARGTDP-SKHKQFRKGIEQLDQEGVVQVLRSDRRGEQAPVLAAVGPMQFE
MSMEG_1316|M.smegmatis_MC2_155 ARGTDP-SKHKQFRRGIEQLEQEGVVQVLRSDKRGEQAPVFAAVGPMQFE
* ... :.:::: .*::* :*. . :: * . *: *:..:* ::::
MMAR_1013|M.marinum_M ILVDRMRREFKVEANVGKPQVAYKETIKRLVEKVEFTHKKQTGGSGQFAK
MUL_0765|M.ulcerans_Agy99 ILVDRMRREFKVEANVGKPQVAYKETIKRLVEKVEFTHKKQTGGSGQFAK
MAV_4490|M.avium_104 ILVDRMRREFKVEANVGKPQVAYKETIRRKVENVEYTHKKQTGGSGQFAK
MLBr_01878|M.leprae_Br4923 ILVDRMRREFKVEANVGKPQVAYKETIRRVVETVEYTHKKQTGGSGQFAK
Mb0703|M.bovis_AF2122/97 ILVDRMRREFKVEANVGKPQVAYKETIKRLVQNVEYTHKKQTGGSGQFAK
Rv0684|M.tuberculosis_H37Rv ILVDRMRREFKVEANVGKPQVAYKETIKRLVQNVEYTHKKQTGGSGQFAK
MAB_3849c|M.abscessus_ATCC_199 ILVDRMRREFKVEANVGKPQVAYRETIRKKVENVEFTHKKQTGGSGQFAK
TH_1890|M.thermoresistible__bu VLVDRMKREFKVEANVGKPQVAYRETIKRKVENVEFTHKKQTGGSGQFAK
Mflv_1328|M.gilvum_PYR-GCK VAAHRMAAEFSAP-------------------------------------
Mvan_5464|M.vanbaalenii_PYR-1 VASHRMAAEFSAP-------------------------------------
MSMEG_1316|M.smegmatis_MC2_155 VASHRMATELSAP-------------------------------------
: .** *:..
MMAR_1013|M.marinum_M VLISIEPFT-GEDGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYGVLA
MUL_0765|M.ulcerans_Agy99 VLISIEPFT-GEDGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYGVLA
MAV_4490|M.avium_104 VIINLEPFT-GEDGATYEFENKVTGGRIPREYIPSVDAGAQDAMQYGVLA
MLBr_01878|M.leprae_Br4923 VIIKLEPFS-GENGATYEFENKVTGGRIPREYIPSVEAGARDAMQYGVLA
Mb0703|M.bovis_AF2122/97 VIINLEPFT-GEEGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYGVLA
Rv0684|M.tuberculosis_H37Rv VIINLEPFT-GEEGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYGVLA
MAB_3849c|M.abscessus_ATCC_199 VIVTVEPLVDAEDGATYEFENKVTGGRVPREYIPSVDAGAQDAMQYGILA
TH_1890|M.thermoresistible__bu VIITIEPFT-GEDGATYEFENKVTGGRIPREYIPSVDAGVQDAMQYGVLA
Mflv_1328|M.gilvum_PYR-GCK --IELESLP-----------------------------------------
Mvan_5464|M.vanbaalenii_PYR-1 --IALESLP-----------------------------------------
MSMEG_1316|M.smegmatis_MC2_155 --ISLEPLP-----------------------------------------
: :*.:
MMAR_1013|M.marinum_M GYPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAAAHPVILEPIM
MUL_0765|M.ulcerans_Agy99 GYPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAAAHPVILEPIM
MAV_4490|M.avium_104 GYPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAQAQPVILEPIM
MLBr_01878|M.leprae_Br4923 GYPLVNLKVTLLDGAYHDVDSSEIAFKIAGSQVLKKAAAQAQPVILEPIM
Mb0703|M.bovis_AF2122/97 GYPLVNLKVTLLDGAYHEVDSSEMAFKIAGSQVLKKAAALAQPVILEPIM
Rv0684|M.tuberculosis_H37Rv GYPLVNLKVTLLDGAYHEVDSSEMAFKIAGSQVLKKAAALAQPVILEPIM
MAB_3849c|M.abscessus_ATCC_199 GYPLVNIKVTLLDGAYHDVDSSEMAFKIAGSQALKKAAQAAQPVILEPLM
TH_1890|M.thermoresistible__bu GYPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQALKKAAQMANPVILEPIM
Mflv_1328|M.gilvum_PYR-GCK -------------------------------YQVARICDPADAEALQKMA
Mvan_5464|M.vanbaalenii_PYR-1 -------------------------------YTIARICDPADAEALAKMA
MSMEG_1316|M.smegmatis_MC2_155 -------------------------------YQVARIVDPEDADFMNRQP
: : .. :
MMAR_1013|M.marinum_M AVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGARVVKAHVPLSEMFGYV
MUL_0765|M.ulcerans_Agy99 AVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGARVVKAHVPLSEMFGYV
MAV_4490|M.avium_104 AVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGARVVKAHVPLSEMFGYV
MLBr_01878|M.leprae_Br4923 AVEVTTPEDYMGDVIGDLHSRRGQIQAMKERAGTRVVRAHVPLSEMFGYV
Mb0703|M.bovis_AF2122/97 AVEVTTPEDYMGDVIGDLNSRRGQIQAMEERAGARVVRAHVPLSEMFGYV
Rv0684|M.tuberculosis_H37Rv AVEVTTPEDYMGDVIGDLNSRRGQIQAMEERAGARVVRAHVPLSEMFGYV
MAB_3849c|M.abscessus_ATCC_199 AVEVITPEDYMGDVIGDLNSRRGQIQAMEERSGARVVKAQVPLSEMFGYV
TH_1890|M.thermoresistible__bu AVEVTTPEEYMGDVIGDLNSRRGQIQAMEERGGARVVKALVPLSEMFGYV
Mflv_1328|M.gilvum_PYR-GCK AVEVFTRTD---GVMLAVFSTKWRLETVQRDNPDIMLRELIAAGQE----
Mvan_5464|M.vanbaalenii_PYR-1 AVEVFTRTD---GVMLAVFSTKWRLETVQRENPAMELRELVAAGQ-----
MSMEG_1316|M.smegmatis_MC2_155 SAEVMTRSD---GVMLVLFSTPWRLEGFQRDNPDIKLRSLVAS-------
:.** * : .*: : * ::: .:. :: :.
MMAR_1013|M.marinum_M GDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
MUL_0765|M.ulcerans_Agy99 GDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
MAV_4490|M.avium_104 GDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGQ
MLBr_01878|M.leprae_Br4923 GDLRSKTQGRANYSMVFNSYSEVPANVSKEIIAKATGE
Mb0703|M.bovis_AF2122/97 GDLRSKTQGRANYSMVFDSYSEVPANVSKEIIAKATGE
Rv0684|M.tuberculosis_H37Rv GDLRSKTQGRANYSMVFDSYSEVPANVSKEIIAKATGE
MAB_3849c|M.abscessus_ATCC_199 GDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
TH_1890|M.thermoresistible__bu GDLRSKSQGRANYSMVFDSYAEVPAQVSKEIIAKATGE
Mflv_1328|M.gilvum_PYR-GCK --------------------------------------
Mvan_5464|M.vanbaalenii_PYR-1 --------------------------------------
MSMEG_1316|M.smegmatis_MC2_155 --------------------------------------