For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAQKDVLTDLTKVRNIGIMAHIDAGKTTTTERILYYTGISYKIGEVHDGAATMDWMEQEQERGITITSAA TTCFWNDNQINIIDTPGHVDFTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM DKIGADFYFSVRTMEERLGANVIPIQLPVGSEGDFEGVVDLVEMKAKVWSADAKLGEKYDVVDIPADLQE KADEYRTKLLEAVAETDEALLEKYLGGEELTEAEIKGAIRKLTITSEAYPVLCGSAFKNKGVQPMLDAVI DYLPSPLDVPAAIGHVPGKEDEEVVRKPSTDEPFSALAFKVATHPFFGKLTYVRVYSGKVDSGSQVINST KGKKERLGKLFQMHSNKESPVETASAGHIYAVIGLKDTTTGDTLSDPNNQIVLESMTFPDPVIEVAIEPK TKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQTVIGGMGELHLDILVDRMRREFKVEANVGKPQVAYKET IKRLVEKVEFTHKKQTGGSGQFAKVLISIEPFTGEDGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYG VLAGYPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAAAHPVILEPIMAVEVTTPEDYMGDVIGD LNSRRGQIQAMEERSGARVVKAHVPLSEMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATG E
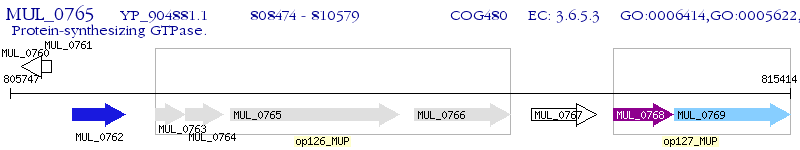
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_0765 | fusA1 | - | 100% (701) | elongation factor G |
| M. ulcerans Agy99 | MUL_1009 | typA | 2e-35 | 27.98% (511) | GTP-binding translation elongation factor TypA |
| M. ulcerans Agy99 | MUL_3667 | lepA | 2e-12 | 33.55% (155) | GTP-binding protein LepA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0703 | fusA1 | 0.0 | 91.30% (701) | elongation factor G |
| M. gilvum PYR-GCK | Mflv_5076 | - | 0.0 | 89.11% (698) | elongation factor G |
| M. tuberculosis H37Rv | Rv0684 | fusA1 | 0.0 | 91.30% (701) | elongation factor G |
| M. leprae Br4923 | MLBr_01878 | fusA | 0.0 | 93.87% (701) | elongation factor G |
| M. abscessus ATCC 19977 | MAB_3849c | - | 0.0 | 85.98% (699) | elongation factor G (EF-G) |
| M. marinum M | MMAR_1013 | fusA1 | 0.0 | 99.86% (701) | elongation factor G, FusA1 |
| M. avium 104 | MAV_4490 | fusA | 0.0 | 96.29% (701) | elongation factor G |
| M. smegmatis MC2 155 | MSMEG_6535 | - | 1e-102 | 31.48% (721) | elongation factor G |
| M. thermoresistible (build 8) | TH_1890 | fusA | 0.0 | 87.87% (701) | PROBABLE ELONGATION FACTOR G FUSA1 (EF-G) |
| M. vanbaalenii PYR-1 | Mvan_1281 | - | 0.0 | 91.83% (698) | elongation factor G |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5076|M.gilvum_PYR-GCK -----------MAQDVLTDLSKVRNIGIMAHIDAGKTTTTERILYYTGVN
MAB_3849c|M.abscessus_ATCC_199 -----------MAQDVLTDLNKVRNIGIMAHIDAGKTTTTERILYYTGVN
TH_1890|M.thermoresistible__bu ----------VAQKDVLTDLRRVRNIGIMAHIDAGKTTTTERILFYTGVN
Mb0703|M.bovis_AF2122/97 ----------MAQKDVLTDLSRVRNFGIMAHIDAGKTTTTERILYYTGIN
Rv0684|M.tuberculosis_H37Rv ----------MAQKDVLTDLSRVRNFGIMAHIDAGKTTTTERILYYTGIN
MUL_0765|M.ulcerans_Agy99 ----------MAQKDVLTDLTKVRNIGIMAHIDAGKTTTTERILYYTGIS
MMAR_1013|M.marinum_M ----------MAQKDVLTDLTKVRNIGIMAHIDAGKTTTTERILYYTGIS
MAV_4490|M.avium_104 ----------MAQKDVLTDLTKVRNIGIMAHIDAGKTTTTERILYYTGIS
MLBr_01878|M.leprae_Br4923 ----------MAQKDVLTDLTKVRNIGIMAHIDAGKTTTTERILYYTGIS
Mvan_1281|M.vanbaalenii_PYR-1 ----------MAQ-DVLTDLTKVRNIGIMAHIDAGKTTTTERILFYTGIS
MSMEG_6535|M.smegmatis_MC2_155 MADRTHSPAGNGSVPTAERPAAIRNVALVGPSGGGKTTLVEALLVAGGVL
. :**..::. ..**** .* :* *:
Mflv_5076|M.gilvum_PYR-GCK YKIGETHDGASTTDWMEQEQERGITITSAAVTCFWNQNQINIIDTPGHVD
MAB_3849c|M.abscessus_ATCC_199 YKIGETHDGASTTDWMEQEQERGITITSAAVTCFWNGNQINIIDTPGHVD
TH_1890|M.thermoresistible__bu YKMGETHDGASTTDWMEQEQERGITITSAAVTCFWNDHQINIIDTPGHVD
Mb0703|M.bovis_AF2122/97 YKIGEVHDGAATMDWMEQEQERGITITSAATTTFWKDNQLNIIDTPGHVD
Rv0684|M.tuberculosis_H37Rv YKIGEVHDGAATMDWMEQEQERGITITSAATTTFWKDNQLNIIDTPGHVD
MUL_0765|M.ulcerans_Agy99 YKIGEVHDGAATMDWMEQEQERGITITSAATTCFWNDNQINIIDTPGHVD
MMAR_1013|M.marinum_M YKIGEVHDGAATMDWMEQEQERGITITSAATTCFWNDNQINIIDTPGHVD
MAV_4490|M.avium_104 YKIGEVHDGAATMDWMEQEQERGITITSAATTCFWNDNQINIIDTPGHVD
MLBr_01878|M.leprae_Br4923 YKIGEVHDGAATMDWMEQEQERGITITSAATTCFWNDNQINIIDTPGHVD
Mvan_1281|M.vanbaalenii_PYR-1 YKIGEVHDGAATMDWMEQEQERGITITSAATTCFWNDNQINIIDTPGHVD
MSMEG_6535|M.smegmatis_MC2_155 TRPGSVADGSTVCDFDEAEIAQQRSVSLALASLSHNGIKVNLIDTPGYAD
: *.. **::. *: * * : ::: * .: : ::*:*****:.*
Mflv_5076|M.gilvum_PYR-GCK FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MAB_3849c|M.abscessus_ATCC_199 FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
TH_1890|M.thermoresistible__bu FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
Mb0703|M.bovis_AF2122/97 FTVEVERNLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
Rv0684|M.tuberculosis_H37Rv FTVEVERNLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MUL_0765|M.ulcerans_Agy99 FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MMAR_1013|M.marinum_M FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MAV_4490|M.avium_104 FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MLBr_01878|M.leprae_Br4923 FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYEVPRICFVNKM
Mvan_1281|M.vanbaalenii_PYR-1 FTVEVERSLRVLDGAVAVFDGKEGVEPQSEQVWRQADKYDVPRICFVNKM
MSMEG_6535|M.smegmatis_MC2_155 FVGELRAGLRAADCALFVIAANETIDEPTKTLWQECNEVQMPRAVVITKL
*. *:. .**. * *: *: .:* :: :: :*::.:: ::** .:.*:
Mflv_5076|M.gilvum_PYR-GCK DKLGADFYFTVRTIEERLGARPLVIQLPIGAENDFIGIIDLVEMKAKVWR
MAB_3849c|M.abscessus_ATCC_199 DKLGADFYFTVQTIKDRLGAKPLVIQLPIGAENDFEGIIDLVEMNAKVWR
TH_1890|M.thermoresistible__bu DKLGADFYFTVDTIKERLGANPLVIQLPIGAENDFVGVVDLVEMKAKVWR
Mb0703|M.bovis_AF2122/97 DKIGADFYFSVRTMGERLGANAVPIQLPVGAEADFEGVVDLVEMNAKVWR
Rv0684|M.tuberculosis_H37Rv DKIGADFYFSVRTMGERLGANAVPIQLPVGAEADFEGVVDLVEMNAKVWR
MUL_0765|M.ulcerans_Agy99 DKIGADFYFSVRTMEERLGANVIPIQLPVGSEGDFEGVVDLVEMKAKVWS
MMAR_1013|M.marinum_M DKIGADFYFSVRTMEERLGANVIPIQLPVGSEGDFEGVVDLVEMKAKVWS
MAV_4490|M.avium_104 DKIGADFYFSVRTMEERLGANVIPIQIPVGSEGDFEGVVDLVEMKAKVWS
MLBr_01878|M.leprae_Br4923 DKIGADFYFSVRTMQERLGANVIPIQLPVGSEGDFEGVVDLVEMKAKVWS
Mvan_1281|M.vanbaalenii_PYR-1 DKLGADFYFSVQTMKDRLGANVVPIQLPIGSEGDFEGVVDLVEMKAKVWR
MSMEG_6535|M.smegmatis_MC2_155 DHPRANYDAALTAAQEAFGDKVAPLYFPVGDGESCKGVVGLLTRTYYDYS
*: *:: :: : : :* . : :*:* . *::.*: . :
Mflv_5076|M.gilvum_PYR-GCK GETALGEKYEVEEIPADLADKAEEYRTKLIEAVAET--DEALLEKYFGGE
MAB_3849c|M.abscessus_ATCC_199 GETKLGESYETVEIPADLADKAAEYRNELLETVAES--DEALLEKYLGGE
TH_1890|M.thermoresistible__bu GETALGEKYEVEEIPAELADKAEEYRNALLEAVAET--DEDLLEKYLGGE
Mb0703|M.bovis_AF2122/97 GETKLGETYDTVEIPADLAEQAEEYRTKLLEVVAES--DEHLLEKYLGGE
Rv0684|M.tuberculosis_H37Rv GETKLGETYDTVEIPADLAEQAEEYRTKLLEVVAES--DEHLLEKYLGGE
MUL_0765|M.ulcerans_Agy99 ADAKLGEKYDVVDIPADLQEKADEYRTKLLEAVAET--DEALLEKYLGGE
MMAR_1013|M.marinum_M ADAKLGEKYDVVDIPADLQEKADEYRTKLLEAVAET--DEALLEKYLGGE
MAV_4490|M.avium_104 AEAKLGEKYDVVDIPADLQEKAEEYRTKLLEAVAET--DEALLDKYLGGE
MLBr_01878|M.leprae_Br4923 TEAKLGEKYDVVGIPTDLQEKAEEYRTNLLETVAET--DEALLEKYFSGE
Mvan_1281|M.vanbaalenii_PYR-1 GETKLGEKYDTVEIPADLQEKAEEYRTAMIEAVAET--DDELMEKYLGGE
MSMEG_6535|M.smegmatis_MC2_155 GGTHTARPPD-----GSHDAAIAELRGSLIEGVIEESEDETLMERYLGGE
: .. : . * * ::* * * *: *:::*:.**
Mflv_5076|M.gilvum_PYR-GCK ELSIDEIKGAIRKLTVASELYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
MAB_3849c|M.abscessus_ATCC_199 ELSIDEIKAGIRKLTVASELYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
TH_1890|M.thermoresistible__bu ELTVEEIKGAIRKLTVTSQAYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
Mb0703|M.bovis_AF2122/97 ELTVDEIKGAIRKLTIASEIYPVLCGSAFKNKGVQPMLDAVVDYLPSPLD
Rv0684|M.tuberculosis_H37Rv ELTVDEIKGAIRKLTIASEIYPVLCGSAFKNKGVQPMLDAVVDYLPSPLD
MUL_0765|M.ulcerans_Agy99 ELTEAEIKGAIRKLTITSEAYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
MMAR_1013|M.marinum_M ELTEAEIKGAIRKLTITSEAYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
MAV_4490|M.avium_104 ELTIEEIKGAIRKLTISSEAYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
MLBr_01878|M.leprae_Br4923 ELTVAEIKGAIRKLTISSEAYPVLCGSAFKNKGVQPMLDAVIDYLPSPLD
Mvan_1281|M.vanbaalenii_PYR-1 ELTIEEIKSGLRKLTITSAGYPVLCGSAFKNKGVQPMLDAVIDYLPNPLD
MSMEG_6535|M.smegmatis_MC2_155 EIDEAVLIADLEKAVARASFFPVIPVCSQSGVGTLELLDIISRGFPAPPE
*: : . :.* . : :**: .: .. *. :** : :* * :
Mflv_5076|M.gilvum_PYR-GCK VESVTGHVPNKEDEVISRKPSTDEPFSALAFKIAVHPFFGKLTYVRVYSG
MAB_3849c|M.abscessus_ATCC_199 VESVKGHVPGHEDQEIERKPSTDEPFSALAFKIAVHPFFGKLTYVRVYSG
TH_1890|M.thermoresistible__bu VESVKGHVPGHEDQEIARKPSVDEPFSALAFKIAVHPFFGKLTYIRVYSG
Mb0703|M.bovis_AF2122/97 VPPAIGHAPAKEDEEVVRKATTDEPFAALAFKIATHPFFGKLTYIRVYSG
Rv0684|M.tuberculosis_H37Rv VPPAIGHAPAKEDEEVVRKATTDEPFAALAFKIATHPFFGKLTYIRVYSG
MUL_0765|M.ulcerans_Agy99 VPAAIGHVPGKEDEEVVRKPSTDEPFSALAFKVATHPFFGKLTYVRVYSG
MMAR_1013|M.marinum_M VPAAIGHVPGKEDEEVVRKPSTDEPFSALAFKVATHPFFGKLTYVRVYSG
MAV_4490|M.avium_104 VPPAEGHVPGKEEELITRKPSTDEPFSALAFKVATHPFFGKLTYVRVYSG
MLBr_01878|M.leprae_Br4923 VPAAIGHVPGKEDEEIVRKPSTDEPLSALAFKVATHPFFGKLTYVRVYSG
Mvan_1281|M.vanbaalenii_PYR-1 VPAAEGHVPGKEDEIISRKPSADEPFSGLAFKVATHPFFGKLTYVRVYSG
MSMEG_6535|M.smegmatis_MC2_155 HQLPEVFTP-QGAPRKPLSCDPDGPLLAEVVKTTSDPYVGRVSLVRVFSG
..* : . * *: . ..* : .*:.*::: :**:**
Mflv_5076|M.gilvum_PYR-GCK TVE---------------------------SGSQVINSTKGKKERLGKLF
MAB_3849c|M.abscessus_ATCC_199 KIE---------------------------SGAQVVNATKGKKERLGKLF
TH_1890|M.thermoresistible__bu QVE---------------------------SGSQVMNSTKGKKERLGKLF
Mb0703|M.bovis_AF2122/97 TVE---------------------------SGSQVINATKGKKERLGKLF
Rv0684|M.tuberculosis_H37Rv TVE---------------------------SGSQVINATKGKKERLGKLF
MUL_0765|M.ulcerans_Agy99 KVD---------------------------SGSQVINSTKGKKERLGKLF
MMAR_1013|M.marinum_M KVD---------------------------SGSQVINSTKGKKERLGKLF
MAV_4490|M.avium_104 KVD---------------------------SGSQVINSTKGKKERLGKLF
MLBr_01878|M.leprae_Br4923 KVD---------------------------SGSQVINATKGKKERLGKLF
Mvan_1281|M.vanbaalenii_PYR-1 KVD---------------------------SGSQVVNSTKGKKERLGKLF
MSMEG_6535|M.smegmatis_MC2_155 TIRPDATVHVSGHFSAFTDTSGSHGAAAAADGSTHSHADHDEDERIGTLS
: .*: :: :.:.**:*.*
Mflv_5076|M.gilvum_PYR-GCK QMHANKENPVERASAGHIYAVIGLKDTTTGDTLCDANQQIVLESMTFPDP
MAB_3849c|M.abscessus_ATCC_199 QMHANKENPVETAAAGHIYAVIGLKDTTTGDTLCDPNSQIVLESMTFPDP
TH_1890|M.thermoresistible__bu QMHSNKENPVERVSAGHIYAVIGLKDTTTGDTLCDPNDQIVLESMTFPDP
Mb0703|M.bovis_AF2122/97 QMHSNKENPVDRASAGHIYAVIGLKDTTTGDTLSDPNQQIVLESMTFPDP
Rv0684|M.tuberculosis_H37Rv QMHSNKENPVDRASAGHIYAVIGLKDTTTGDTLSDPNQQIVLESMTFPDP
MUL_0765|M.ulcerans_Agy99 QMHSNKESPVETASAGHIYAVIGLKDTTTGDTLSDPNNQIVLESMTFPDP
MMAR_1013|M.marinum_M QMHSNKENPVETASAGHIYAVIGLKDTTTGDTLSDPNNQIVLESMTFPDP
MAV_4490|M.avium_104 QMHSNKENPVETASAGHIYAVIGLKDTTTGDTLSDPNHQIVLESMTFPDP
MLBr_01878|M.leprae_Br4923 QMHSNKENPVETASAGHIYAVIGLKDTTTGDTLADPNNQIVLESMTFPDP
Mvan_1281|M.vanbaalenii_PYR-1 QMHSNKENPVESASAGHIYAVIGLKDTTTGDTLCDPNQQIVLESMTFPDP
MSMEG_6535|M.smegmatis_MC2_155 FPLGKQQRPAPTVVAGDICAIGRLSRAETGDTLSDKTDPLVLRPWTMPDP
.::: *. . **.* *: *. : *****.* . :**.. *:***
Mflv_5076|M.gilvum_PYR-GCK VIEVAIEPKTKSDQEKLGTAIQKLAEEDPTFKVHLDQETGQTVIGGMGEL
MAB_3849c|M.abscessus_ATCC_199 VIEVAIEPKTKTDQEKLGTAIQKLAEEDPTFKVKLDQETGQTVIGGMGEL
TH_1890|M.thermoresistible__bu VIEVAIEPKTKSDQEKLGTAIQKLAEEDPTFKVHQDPETGQTVIGGMGEL
Mb0703|M.bovis_AF2122/97 VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQTVIGGMGEL
Rv0684|M.tuberculosis_H37Rv VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQTVIGGMGEL
MUL_0765|M.ulcerans_Agy99 VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQTVIGGMGEL
MMAR_1013|M.marinum_M VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQTVIGGMGEL
MAV_4490|M.avium_104 VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDQETGQTVIGGMGEL
MLBr_01878|M.leprae_Br4923 VIEVAIEPKTKSDQEKLSLSIQKLAEEDPTFKVHLDSETGQTVIGGMGEL
Mvan_1281|M.vanbaalenii_PYR-1 VIEVAIEPKTKSDQEKLGTAIQKLAEEDPTFKVHLDQETGQTVIGGMGEL
MSMEG_6535|M.smegmatis_MC2_155 LLPIAVAPRAKTDEDKLSVGLQRLAAEDPTLRIEQNPETHQIVLWCMGEA
:: :*: *::*:*::**. .:*:** ****:::. : ** * *: ***
Mflv_5076|M.gilvum_PYR-GCK HLDILVDRMRREFKVEANVGKPQVAYRETIRRKVEKVEFTHKKQTGGSGQ
MAB_3849c|M.abscessus_ATCC_199 HLDILVDRMRREFKVEANVGKPQVAYRETIRKKVENVEFTHKKQTGGSGQ
TH_1890|M.thermoresistible__bu HLDVLVDRMKREFKVEANVGKPQVAYRETIKRKVENVEFTHKKQTGGSGQ
Mb0703|M.bovis_AF2122/97 HLDILVDRMRREFKVEANVGKPQVAYKETIKRLVQNVEYTHKKQTGGSGQ
Rv0684|M.tuberculosis_H37Rv HLDILVDRMRREFKVEANVGKPQVAYKETIKRLVQNVEYTHKKQTGGSGQ
MUL_0765|M.ulcerans_Agy99 HLDILVDRMRREFKVEANVGKPQVAYKETIKRLVEKVEFTHKKQTGGSGQ
MMAR_1013|M.marinum_M HLDILVDRMRREFKVEANVGKPQVAYKETIKRLVEKVEFTHKKQTGGSGQ
MAV_4490|M.avium_104 HLDILVDRMRREFKVEANVGKPQVAYKETIRRKVENVEYTHKKQTGGSGQ
MLBr_01878|M.leprae_Br4923 HLDILVDRMRREFKVEANVGKPQVAYKETIRRVVETVEYTHKKQTGGSGQ
Mvan_1281|M.vanbaalenii_PYR-1 HLDILVDRMRREFKVEANVGKPQVAYRETIRRKVEKVEYTHKKQTGGSGQ
MSMEG_6535|M.smegmatis_MC2_155 HAAVVLDALSRRYGVSVDTVELRVPLRETFAGKATGHG-RHVKQSGGHGQ
* :::* : *.: *..:. : :*. :**: . * **:** **
Mflv_5076|M.gilvum_PYR-GCK FAKVLIDLEPFS-GEDGATYEFENKVTGGRIPREYIPSVDAGAQDAMQYG
MAB_3849c|M.abscessus_ATCC_199 FAKVIVTVEPLVDAEDGATYEFENKVTGGRVPREYIPSVDAGAQDAMQYG
TH_1890|M.thermoresistible__bu FAKVIITIEPFT-GEDGATYEFENKVTGGRIPREYIPSVDAGVQDAMQYG
Mb0703|M.bovis_AF2122/97 FAKVIINLEPFT-GEEGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYG
Rv0684|M.tuberculosis_H37Rv FAKVIINLEPFT-GEEGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYG
MUL_0765|M.ulcerans_Agy99 FAKVLISIEPFT-GEDGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYG
MMAR_1013|M.marinum_M FAKVLISIEPFT-GEDGATYEFESKVTGGRIPREYIPSVDAGAQDAMQYG
MAV_4490|M.avium_104 FAKVIINLEPFT-GEDGATYEFENKVTGGRIPREYIPSVDAGAQDAMQYG
MLBr_01878|M.leprae_Br4923 FAKVIIKLEPFS-GENGATYEFENKVTGGRIPREYIPSVEAGARDAMQYG
Mvan_1281|M.vanbaalenii_PYR-1 FAKVLIDLEPFT-GEDGATYEFENKVTGGRIPREYIPSVDAGAQDAMQYG
MSMEG_6535|M.smegmatis_MC2_155 YAVCDIEVEPLP---EGSGFEFVDKVVGGAVPRQFIPSVEKGVRAQMEKG
:* : :**: :*: :** .**.** :**::****: *.: *: *
Mflv_5076|M.gilvum_PYR-GCK VLAG--YPLVNIKVTLLDGAFHEVDSSEMAFKVAGSQVLKKAAQAAQPVI
MAB_3849c|M.abscessus_ATCC_199 ILAG--YPLVNIKVTLLDGAYHDVDSSEMAFKIAGSQALKKAAQAAQPVI
TH_1890|M.thermoresistible__bu VLAG--YPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQALKKAAQMANPVI
Mb0703|M.bovis_AF2122/97 VLAG--YPLVNLKVTLLDGAYHEVDSSEMAFKIAGSQVLKKAAALAQPVI
Rv0684|M.tuberculosis_H37Rv VLAG--YPLVNLKVTLLDGAYHEVDSSEMAFKIAGSQVLKKAAALAQPVI
MUL_0765|M.ulcerans_Agy99 VLAG--YPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAAAHPVI
MMAR_1013|M.marinum_M VLAG--YPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAAAHPVI
MAV_4490|M.avium_104 VLAG--YPLVNLKVTLLDGAFHEVDSSEMAFKIAGSQVLKKAAAQAQPVI
MLBr_01878|M.leprae_Br4923 VLAG--YPLVNLKVTLLDGAYHDVDSSEIAFKIAGSQVLKKAAAQAQPVI
Mvan_1281|M.vanbaalenii_PYR-1 VLAG--YPLVNVKVTLLDGAFHEVDSSEMAFKVAGSQVLKKAAQSAQPVI
MSMEG_6535|M.smegmatis_MC2_155 VTGESGYPVVDIRVTLFDGKAHSVDSSDFAFQMAGALALRDAAAATKINL
: . **:*:::***:** *.****::**::**: .*:.** :: :
Mflv_5076|M.gilvum_PYR-GCK LEPIMAVEVITPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAQVPLS
MAB_3849c|M.abscessus_ATCC_199 LEPLMAVEVITPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAQVPLS
TH_1890|M.thermoresistible__bu LEPIMAVEVTTPEEYMGDVIGDLNSRRGQIQAMEERGGAR-VVKALVPLS
Mb0703|M.bovis_AF2122/97 LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERAGAR-VVRAHVPLS
Rv0684|M.tuberculosis_H37Rv LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERAGAR-VVRAHVPLS
MUL_0765|M.ulcerans_Agy99 LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAHVPLS
MMAR_1013|M.marinum_M LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAHVPLS
MAV_4490|M.avium_104 LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAHVPLS
MLBr_01878|M.leprae_Br4923 LEPIMAVEVTTPEDYMGDVIGDLHSRRGQIQAMKERAGTR-VVRAHVPLS
Mvan_1281|M.vanbaalenii_PYR-1 LEPIMAVEVTTPEDYMGDVIGDLNSRRGQIQAMEERSGAR-VVKAQVPLS
MSMEG_6535|M.smegmatis_MC2_155 LEPVDDVTIVVPDDLVGTIMGDLSGRRGRVLGTDKHDADRTVIKAEIPEV
***: * : .*:: :* ::*** .***:: . .:: . * *::* :*
Mflv_5076|M.gilvum_PYR-GCK EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGQ
MAB_3849c|M.abscessus_ATCC_199 EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
TH_1890|M.thermoresistible__bu EMFGYVGDLRSKSQGRANYSMVFDSYAEVPAQVSKEIIAKATGE
Mb0703|M.bovis_AF2122/97 EMFGYVGDLRSKTQGRANYSMVFDSYSEVPANVSKEIIAKATGE
Rv0684|M.tuberculosis_H37Rv EMFGYVGDLRSKTQGRANYSMVFDSYSEVPANVSKEIIAKATGE
MUL_0765|M.ulcerans_Agy99 EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
MMAR_1013|M.marinum_M EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGE
MAV_4490|M.avium_104 EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGQ
MLBr_01878|M.leprae_Br4923 EMFGYVGDLRSKTQGRANYSMVFNSYSEVPANVSKEIIAKATGE
Mvan_1281|M.vanbaalenii_PYR-1 EMFGYVGDLRSKTQGRANYSMVFDSYAEVPANVSKEIIAKATGQ
MSMEG_6535|M.smegmatis_MC2_155 ELVRYAIDLRSMSHGAGQFRRSFARYEPMPESAAARLRTSA---
*:. *. **** ::* .:: * * :* ..: .: :.*