For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FAQLGIERVDYKDTSTLRQFLSERGKIRSRTVTGLTVQQQRQVTIAVKNAREMALLPYPGQG*
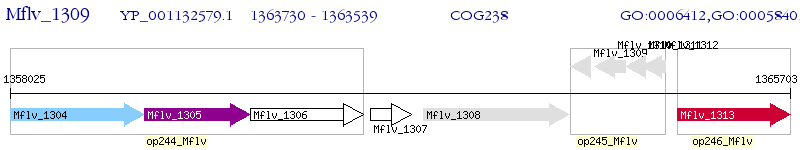
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1309 | - | - | 100% (63) | ribosomal protein S18 |
| M. gilvum PYR-GCK | Mflv_0876 | rpsR | 7e-15 | 68.00% (50) | 30S ribosomal protein S18 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2081c | rpsR | 7e-23 | 82.76% (58) | 30S ribosomal protein S18 |
| M. tuberculosis H37Rv | Rv2055c | rpsR | 7e-23 | 82.76% (58) | 30S ribosomal protein S18 |
| M. leprae Br4923 | MLBr_02683 | rpsR | 7e-16 | 65.38% (52) | 30S ribosomal protein S18 |
| M. abscessus ATCC 19977 | MAB_0331c | rpsR.2 | 1e-22 | 81.36% (59) | 30S ribosomal protein S18 |
| M. marinum M | MMAR_0289 | rpsR2 | 3e-20 | 76.79% (56) | ribosomal protein S18 RpsR2 |
| M. avium 104 | MAV_0076 | rpsR | 4e-15 | 65.38% (52) | 30S ribosomal protein S18 |
| M. smegmatis MC2 155 | MSMEG_6065 | rpsR | 2e-18 | 73.68% (57) | ribosomal protein S18 |
| M. thermoresistible (build 8) | TH_2094 | rpsR | 5e-14 | 66.00% (50) | PROBABLE 30S RIBOSOMAL PROTEIN S18-1 RPSR1 |
| M. ulcerans Agy99 | MUL_4834 | rpsR2 | 2e-20 | 76.79% (56) | 30S ribosomal protein S18 |
| M. vanbaalenii PYR-1 | Mvan_5492 | - | 1e-25 | 87.10% (62) | 30S ribosomal protein S18 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1309|M.gilvum_PYR-GCK -----------------------MFAQLGIERVDYKDTSTLRQFLSERGK
Mvan_5492|M.vanbaalenii_PYR-1 -MTARRRE------AAPAKKRRNLLKSLGIERVDYKDTSTLRQFISERGK
Mb2081c|M.bovis_AF2122/97 -MAAKSAR------KGPTKAKKNLLDSLGVESVDYKDTATLRVFISDRGK
Rv2055c|M.tuberculosis_H37Rv -MAAKSAR------KGPTKAKKNLLDSLGVESVDYKDTATLRVFISDRGK
MMAR_0289|M.marinum_M -MKKKPRRTRRPIAAPATPAKKNLLDSLGVSEVDYKDISRLRTFISDRGK
MUL_4834|M.ulcerans_Agy99 -MKKKPRRTRRPIAAPATPAKKNLLDSLGVSEVDYKDISRLRTFISDRGK
MAB_0331c|M.abscessus_ATCC_199 -MAPKTPRNRRAKLVDTKPAKPNLFAKLGVSSVDYKDTNTLRMFLSERGK
MSMEG_6065|M.smegmatis_MC2_155 MMAVKKSR-KRTAATELKKPRRNQLEALGVTTIDYKDVAVLRTFLSERGK
MLBr_02683|M.leprae_Br4923 -MAKSTKR--RPAPEKPAKARKCVFCAKKNQQIDYKDTTLLRTYISERGK
MAV_0076|M.avium_104 -MAKSNKR--RPAPEKPVKARKCVFCAKKDQAIDYKDTALLRTYISERGK
TH_2094|M.thermoresistible__bu -MAKSNKR--RPAPEKPVKTRKCVFCSKKGQVIDYKDTALLRTYISERGK
: :**** ** ::*:***
Mflv_1309|M.gilvum_PYR-GCK IRSRTVTGLTVQQQRQVTIAVKNAREMALLPYPGQG---------
Mvan_5492|M.vanbaalenii_PYR-1 IRSRSVTGLTVQQQRQVTTAVKTAREMALLPYPGQNPTGR-----
Mb2081c|M.bovis_AF2122/97 IRSRGVTGLTVQQQRQVAQAIKNAREMALLPYPGQDRQRRAALCP
Rv2055c|M.tuberculosis_H37Rv IRSRGVTGLTVQQQRQVAQAIKNAREMALLPYPGQDRQRRAALCP
MMAR_0289|M.marinum_M IRSRRVTGLTVQQQRQIATAIKNAREMALLPYPASRPQ-------
MUL_4834|M.ulcerans_Agy99 IRSRRVTGLTVQQQRQIATAIKNAREMALLPYPASRPQ-------
MAB_0331c|M.abscessus_ATCC_199 IRSRRVTGLTVQQQRQVANAIKNAREMALLPYA------------
MSMEG_6065|M.smegmatis_MC2_155 IRSRHVTGLTPQQQRQVATAIKNAREMALLPMAGPR---------
MLBr_02683|M.leprae_Br4923 IRARRVTGNCVQHQRDIAIAVKNAREVALLPFTSSAR--------
MAV_0076|M.avium_104 IRARRVTGNCVQHQRDIAIAVKNAREVALLPFTSSAR--------
TH_2094|M.thermoresistible__bu IRARRVTGNCVQHQSDIANAVKNAREMALLPFTSSAR--------
**:* *** *:* ::: *:*.***:**** .