For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GNYEAVTVEIADHIAQVTLIGPGKGNAMGPAFWSELPVVFRELDADPDVRAIVLVGSGRNFSYGLDLAAM GDTLGSMMSDASSSKPRADFHTRLKSMQDAITAVADCRTPTIASVHGWCIGGGVDLISAVDIRYASSDAK FSVREVKLAIVADVGSLARLPLILSDGHLRELALTGKDIDAARAEKIGLVNDVHPDADASLAAARATAAE IAANPPHTVHGIKDVLDEQRTAQVAASLRYVAAWNSAFLPSKDLTEGITAMFEKRAPNFTGE*
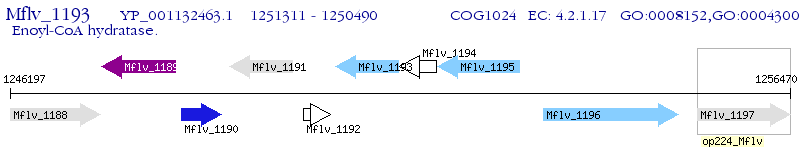
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1193 | - | - | 100% (273) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 6e-33 | 34.70% (268) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_4354 | - | 4e-25 | 31.72% (268) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3803 | echA21 | 1e-118 | 77.04% (270) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv3774 | echA21 | 1e-118 | 77.04% (270) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_00120 | echA1 | 1e-115 | 74.07% (270) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3350 | - | 1e-102 | 67.29% (269) | enoyl-CoA hydratase |
| M. marinum M | MMAR_5329 | echA21 | 1e-118 | 76.67% (270) | enoyl-CoA hydratase, EchA21 |
| M. avium 104 | MAV_0247 | - | 1e-121 | 80.74% (270) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_6353 | - | 1e-124 | 83.33% (270) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_1663 | echA21 | 1e-120 | 79.26% (270) | POSSIBLE ENOYL-CoA HYDRATASE ECHA21 (ENOYL HYDRASE) |
| M. ulcerans Agy99 | MUL_4410 | - | 1e-118 | 76.30% (270) | fusion of enoyl-CoA hydratase, EchA21 and lipase, LipE |
| M. vanbaalenii PYR-1 | Mvan_5613 | - | 1e-142 | 91.97% (274) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3803|M.bovis_AF2122/97 MG--------------------------------ETYESVTVETKDQVAQ
Rv3774|M.tuberculosis_H37Rv MG--------------------------------ETYESVTVETKDQVAQ
MLBr_00120|M.leprae_Br4923 MGS----------------------------DINRAYESVAVEIKDRVAQ
MMAR_5329|M.marinum_M MG--------------------------------ETYESVTVEIKDQVAQ
MUL_4410|M.ulcerans_Agy99 MG--------------------------------ETYESVTVEIKDQVAQ
MAV_0247|M.avium_104 MA--------------------------------QAYESVTVEKKGHVAQ
Mflv_1193|M.gilvum_PYR-GCK MG--------------------------------N-YEAVTVEIADHIAQ
Mvan_5613|M.vanbaalenii_PYR-1 MG--------------------------------NSYESVTVDIADHIAQ
MSMEG_6353|M.smegmatis_MC2_155 MA--------------------------------DTYESVTVEVKDHVAQ
TH_1663|M.thermoresistible__bu MA--------------------------------DTYESVTVDVDGHVAR
MAB_3350|M.abscessus_ATCC_1997 MADSLHASGSSVSDSLRASGSSVSDSLHASGSSADGWKAFTVETTGHIAQ
*. :::.:*: .::*:
Mb3803|M.bovis_AF2122/97 VTLIGPGKGNAMGPAFWSEMPEVFHALDADREVRAIVITGSGKNFSYGLD
Rv3774|M.tuberculosis_H37Rv VTLIGPGKGNAMGPAFWSEMPEVFHALDADREVRAIVITGSGKNFSYGLD
MLBr_00120|M.leprae_Br4923 VTLIGPGKGNAMGPAFWSEMPEVFAELDTNREVRAVVITGSGKDFSYGLD
MMAR_5329|M.marinum_M VTLIGPGKGNAMGPAFWSELPEVFAALDADRDVRAIVITGSGKNFSYGLD
MUL_4410|M.ulcerans_Agy99 ATLIGPGKGNAMGPAFWSELPEVFAALDADRDVRAIVITGSGKNFSYGLD
MAV_0247|M.avium_104 VTLIGPGKGNAMGPAFWSELPELFETLDADPEVRAIVLTGSGKNFSYGLD
Mflv_1193|M.gilvum_PYR-GCK VTLIGPGKGNAMGPAFWSELPVVFRELDADPDVRAIVLVGSGRNFSYGLD
Mvan_5613|M.vanbaalenii_PYR-1 VMLIGPGKGNAMGPAFWAELPVVFHELDADPEVRAIVLTGSGKNFSYGLD
MSMEG_6353|M.smegmatis_MC2_155 VTLIGPGKGNAMGPAFWEEMPDVFGTLDDDPDVRAIVLTGSGKNFSYGLD
TH_1663|M.thermoresistible__bu VTLIGPGKGNAMGPAFWRELPEVFTRLDEDQNVRAIVLTGSGKNFSYGLD
MAB_3350|M.abscessus_ATCC_1997 VTLIGPGKGNAMGPDFWRELPLIFTELDADPEVRAVVLAAAGRHFSYGLD
. ************ ** *:* :* ** : :***:*:..:*:.******
Mb3803|M.bovis_AF2122/97 VPAMGGMFAPLIADGALARPRTDFHTEILRMQKAINAVADCRTPTIAAVQ
Rv3774|M.tuberculosis_H37Rv VPAMGGMFAPLIADGALARPRTDFHTEILRMQKAINAVADCRTPTIAAVQ
MLBr_00120|M.leprae_Br4923 VPAMGGKFVPLLTDGALARPRTDFHAEVLRMQKAINAVADCRTPTIAAVH
MMAR_5329|M.marinum_M VPAMGGTFAPLMAEGALARPRTDFHAEVLRMQKATNAVADCRTPTIASVH
MUL_4410|M.ulcerans_Agy99 VPAMGGTFAPLMAEGALARPRTDFHAEVLRMQKATNAVADCRTPTIASVH
MAV_0247|M.avium_104 VPAMGGSFTPLLSGDALAGPRAVFHREVKRMQGAITAVADCRTPTIASVH
Mflv_1193|M.gilvum_PYR-GCK LAAMGDTLGSMMSDASSSKPRADFHTRLKSMQDAITAVADCRTPTIASVH
Mvan_5613|M.vanbaalenii_PYR-1 LIAMGDTLGSMMADASTSKPRADFHARLKTMQSAITAVADCRTPTIASVH
MSMEG_6353|M.smegmatis_MC2_155 LPAMGASLPALDAGAKS---RSDFHKRLQKMQGAITAVADCRTPTIASVH
TH_1663|M.thermoresistible__bu LPAMGDSLAGVMGPDASARSRAEFHATVQRMQAAINAVADCRTPTIASVH
MAB_3350|M.abscessus_ATCC_1997 LPAMAGTFMPLMAEKALAKPRTDFLDEIRRLQASVTAVADCRKPVIAAIQ
: **. : : *: * : :* : .******.*.**:::
Mb3803|M.bovis_AF2122/97 GWCIGGAVDLISAVDIRYASADAKFSVREVKLAIVADMGSLARLPLILSD
Rv3774|M.tuberculosis_H37Rv GWCIGGAVDLISAVDIRYASADAKFSVREVKLAIVADMGSLARLPLILSD
MLBr_00120|M.leprae_Br4923 GWCIGGALDLISAVDIRYASADAKFSMREVKLAIVADMGSLARLPLILSD
MMAR_5329|M.marinum_M GWCIGGGVDLISAVDMRYASADAKFSIREVKLAIVADMGSLARLPMILND
MUL_4410|M.ulcerans_Agy99 GWCIGGGVDLISAVDMRYASADAKFSIREVKLAIVADMGSLARLPMILND
MAV_0247|M.avium_104 GWCIGGGVDLISAVDIRYASADAKFSVREVKLAIVADVGSLARLPYILND
Mflv_1193|M.gilvum_PYR-GCK GWCIGGGVDLISAVDIRYASSDAKFSVREVKLAIVADVGSLARLPLILSD
Mvan_5613|M.vanbaalenii_PYR-1 GWCIGGGVDLISAVDIRYASSDAKFSVREVKLAIVADVGSLARLPMILSD
MSMEG_6353|M.smegmatis_MC2_155 GWCIGGGVDLISAVDIRYASADAKFSVREVKLAIVADVGSLARLPLILSD
TH_1663|M.thermoresistible__bu GWCIGGGVDLISAVDIRYASADARFSVREVKLAIVADVGSLARLPLILND
MAB_3350|M.abscessus_ATCC_1997 GWCIGGGVDLIAAADIRYASSEAKFSVREAKVAIVADIGSLHRLPPIIGD
******.:***:*.*:****::*:**:**.*:*****:*** *** *:.*
Mb3803|M.bovis_AF2122/97 GHLRELALTGKNIDAARAEKIGLVNDVYDDADQTLAAAHATAAEIAANPP
Rv3774|M.tuberculosis_H37Rv GHLRELALTGKNIDAARAEKIGLVNDVYDDADQTLAAAHATAAEIAANPP
MLBr_00120|M.leprae_Br4923 GHLRELALTGKDIDATRAEKIGLVNDVYDNADKTLAAAHATAAEIAANPP
MMAR_5329|M.marinum_M GHLRELALTGRDIDAARAEKIGLVNQVFEDADATLAAAHATATEIAANPP
MUL_4410|M.ulcerans_Agy99 GHLRELALTGRDIDAARAEKIGLVNQVFEDADATLAAAHATATEIAANPP
MAV_0247|M.avium_104 GHLRELALTGKDIDAARAEKIGLVNDVYADADACLAAAHATAAEIAANPP
Mflv_1193|M.gilvum_PYR-GCK GHLRELALTGKDIDAARAEKIGLVNDVHPDADASLAAARATAAEIAANPP
Mvan_5613|M.vanbaalenii_PYR-1 GHLRELALTGKDIDAARAEKIGLVNDVYPDAEASLAAARATAAEIAANPP
MSMEG_6353|M.smegmatis_MC2_155 GHLRELALTGKDIDAARAEKIGLVNDVYDDAEASLAAAHATAAEIAANPP
TH_1663|M.thermoresistible__bu GHLRELALTGKDIDAGRAEKIGLVNDVFADAEASLEAAHRTAAEIAANPP
MAB_3350|M.abscessus_ATCC_1997 GHLRELAFTGKDIDAARAEKIGLVNDVFDTPEATLAAAHATAAEIAANPP
*******:**::*** *********:*. .: * **: **:*******
Mb3803|M.bovis_AF2122/97 LAVYGIKDVLDQQRTSAVSENLRYVAAWNAAFLPSKDLTEGISATFAKRP
Rv3774|M.tuberculosis_H37Rv LAVYGIKDVLDQQRTSAVSENLRYVAAWNAAFLPSKDLTEGISATFAKRP
MLBr_00120|M.leprae_Br4923 LAVYGVKDVLYQQRTSAISESLRYVAAWNAAFLPSKDLTEGISATFAKRA
MMAR_5329|M.marinum_M LAVYGVKDVLDQQRASAVAENLRYVAAWNAAFLPSKDLTEGISATFAKRP
MUL_4410|M.ulcerans_Agy99 LAVYGVKDVLDQQRASAVAENLRYVAAWNAAFLPSKDLTEGISATFAKRP
MAV_0247|M.avium_104 LTVHGIKDVLDQQRASAVAESLRYVAAWNAAFLPSKDLTEGIAATFEKRP
Mflv_1193|M.gilvum_PYR-GCK HTVHGIKDVLDEQRTAQVAASLRYVAAWNSAFLPSKDLTEGITAMFEKRA
Mvan_5613|M.vanbaalenii_PYR-1 HTVHGIKDVLDEQRTAQVSASLRYVAAWNSAFLPSKDLTEGIKAMFEKRP
MSMEG_6353|M.smegmatis_MC2_155 LTVAGVKDVLDAQRTAQVAESLRYVAAWNSAFLPSKDLAEAVTAMFEKRR
TH_1663|M.thermoresistible__bu LTVRGIKDVLDQQRIDRVSASLRYVAAWNAAFLPSKDLTEGIAATFEKRA
MAB_3350|M.abscessus_ATCC_1997 LVVQGVKDVLGRERENVVADGLKYVSTWNAAFLPSEDLQEAIGAVFEKRE
.* *:**** :* :: .*:**::**:*****:** *.: * * **
Mb3803|M.bovis_AF2122/97 PQFTGE--------------------------------------------
Rv3774|M.tuberculosis_H37Rv PQFTGE--------------------------------------------
MLBr_00120|M.leprae_Br4923 PQFIGE--------------------------------------------
MMAR_5329|M.marinum_M PQFTGE--------------------------------------------
MUL_4410|M.ulcerans_Agy99 PQFTGEQAAASTYPRGVASMTEDGRIRVPADLDAVTALGAEDHSEIDSAA
MAV_0247|M.avium_104 AQFTGE--------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK PNFTGE--------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 PSFTGE--------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 PEFTGE--------------------------------------------
TH_1663|M.thermoresistible__bu PNFTGE--------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 PDFKGR--------------------------------------------
..* *.
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 IERIWQATRYWYQAGMHPAIQLCIRHNGRVVLNRAIGHGWGNAPSDAPDA
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 EKIPVSTDTPFCTYSSAKAITATVVHMLAERGHFSLDDRVCEYIPSYTSH
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 GKDHTTIRHVLTHSAGVPFPTGPRPDVTRADDHDYAARKLGELRPLYPPG
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 LVHIYHALTWGPLMREIIWATTRKEIRDILATEILDPLGFRWTNFGVAEE
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 DLPLVAPSHATGRPLPPVIAAAFRKAIGGTVHEVIPYTNKPLFLRTIIPS
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 SNTVSTANELSRFMEILRRGGELDGVRIMAPETVRSAVTECRRLRPDSAT
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 --------------------------------------------------
Rv3774|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00120|M.leprae_Br4923 --------------------------------------------------
MMAR_5329|M.marinum_M --------------------------------------------------
MUL_4410|M.ulcerans_Agy99 GLAPLRWGTGFMLGSNRFGPFGRNAPAAFGHLGLVNIATWADPDRGLSVG
MAV_0247|M.avium_104 --------------------------------------------------
Mflv_1193|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6353|M.smegmatis_MC2_155 --------------------------------------------------
TH_1663|M.thermoresistible__bu --------------------------------------------------
MAB_3350|M.abscessus_ATCC_1997 --------------------------------------------------
Mb3803|M.bovis_AF2122/97 ---------------------------------
Rv3774|M.tuberculosis_H37Rv ---------------------------------
MLBr_00120|M.leprae_Br4923 ---------------------------------
MMAR_5329|M.marinum_M ---------------------------------
MUL_4410|M.ulcerans_Agy99 LINSGKPGRDPDAKRYVALKNAITAEIPPKTVE
MAV_0247|M.avium_104 ---------------------------------
Mflv_1193|M.gilvum_PYR-GCK ---------------------------------
Mvan_5613|M.vanbaalenii_PYR-1 ---------------------------------
MSMEG_6353|M.smegmatis_MC2_155 ---------------------------------
TH_1663|M.thermoresistible__bu ---------------------------------
MAB_3350|M.abscessus_ATCC_1997 ---------------------------------