For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTLAFGEWIIHRRWYAGRTRELASAQPAAVTALGDGLDHVLLDVAYTDGFTERYQILVQWESAPVDRYGE AALIGTSSGPDGTRFAYDALFNPEAASRLLRLINSSETVGELTFSREPDVTLPVDAPAKVSGAEQSNTSV IFGKDAMLKVFRRVTPGINPDIELNRVLARAGNPRVATLLGSFETSSCALGMVTAFAANSAEGWDMATAS VHDLFANEVGGDFADESRRLGEAVASVHATLAGALGTSVAEFPIDTVLDRLRVATQAAPELAPYAPRIEE RFRRLAERPIQVHRVHGDLHLGQVLRTPESWLLIDFEGEPGQPLEDRRRPDSPLRDVAGVLRSFEYAAYQ QVVTGGGDAQMAERARSWVDRNVDAFCAGYAAVAGEDPRASGDVLAAYELDKAVYEAAYEARFRPSWLPI PMRSIQRLVS
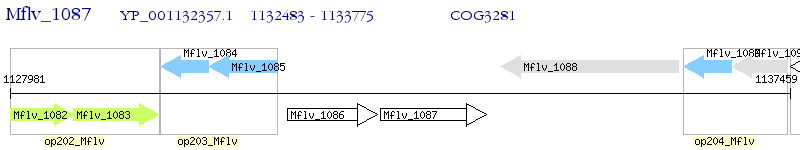
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1087 | - | - | 100% (430) | hypothetical protein Mflv_1087 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0132 | - | 1e-145 | 60.71% (448) | hypothetical protein Mb0132 |
| M. tuberculosis H37Rv | Rv0127 | - | 1e-145 | 60.71% (448) | hypothetical protein Rv0127 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0326 | - | 1e-151 | 62.95% (448) | hypothetical protein MMAR_0326 |
| M. avium 104 | MAV_5185 | - | 1e-147 | 61.92% (449) | trehalose synthase-fused maltokinase |
| M. smegmatis MC2 155 | MSMEG_6514 | - | 1e-152 | 62.61% (444) | trehalose synthase-fused maltokinase |
| M. thermoresistible (build 8) | TH_1781 | - | 1e-152 | 60.34% (464) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4796 | - | 1e-150 | 62.72% (448) | hypothetical protein MUL_4796 |
| M. vanbaalenii PYR-1 | Mvan_5735 | - | 0.0 | 73.64% (440) | hypothetical protein Mvan_5735 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0132|M.bovis_AF2122/97 MTRSDTLATKLPWSDWLPRQRWYAGRNRELATVKPGVVVALRHNLDLVLV
Rv0127|M.tuberculosis_H37Rv MTRSDTLATKLPWSDWLSRQRWYAGRNRELATVKPGVVVALRHNLDLVLV
MMAR_0326|M.marinum_M MNSPATLAAKLPWSEWLPQQRWYAGRNRELAAAEPGAVVALRDDLDLVLV
MUL_4796|M.ulcerans_Agy99 MNSPATLAAKLPWSEWLPQQRWYAGRNRELAAAEPGAVVALRDDLDLVLV
MAV_5185|M.avium_104 MTEP----AKLPWSDWLPQQRWYAGRNRRLTGAEPSVIVGLRDDLDLVLV
MSMEG_6514|M.smegmatis_MC2_155 --------MSVEFEDWLTQQRWYAGRNRELVSATTAMAVRLRDGLELVLL
TH_1781|M.thermoresistible__bu --------MNLPFKDWLPHQRWYAGRSRELVTAEPATVIALSEDMDLVLL
Mflv_1087|M.gilvum_PYR-GCK --------MTLAFGEWIIHRRWYAGRTRELASAQPAAVTALGDGLDHVLL
Mvan_5735|M.vanbaalenii_PYR-1 --------MTLAFGDWIVHRRWYAGRSRELVSAEPAVVTPLRDDLDHILL
.: : :*: ::******.*.*. . .. * ..:: :*:
Mb0132|M.bovis_AF2122/97 DVTYTDGATERYQVLVGWD-------------FEPASEYGTKAAIGVA--
Rv0127|M.tuberculosis_H37Rv DVTYTDGATERYQVLVGWD-------------FEPASEYGTKAAIGVA--
MMAR_0326|M.marinum_M DVSYTDGSAERYQVLVRWD-------------AGPVSEFSTLATIGSA--
MUL_4796|M.ulcerans_Agy99 DVSYTDGSAERYQVLVRWD-------------AGPVSEFSTLATIGSA--
MAV_5185|M.avium_104 DADYADGSRDRYQVLVCWD-------------AAPVSEYSTVATIGAA--
MSMEG_6514|M.smegmatis_MC2_155 QANYADGPDERYQVIV-------------ATGSGPIDEYSVVATIGIA--
TH_1781|M.thermoresistible__bu DVDYRDGSAEKYQIIVSWHADGPQGGPDRGHGGGPIEEYSAVATIGSA--
Mflv_1087|M.gilvum_PYR-GCK DVAYTDGFTERYQILVQWE-------------SAPVDRYGEAALIGTSSG
Mvan_5735|M.vanbaalenii_PYR-1 DVTYTDGTVERYQLVVRWA-------------DSPVAGFGEAATIGTALG
:. * ** ::**::* * :. * ** :
Mb0132|M.bovis_AF2122/97 --DDRTGFDALYDVAGPQFLLSLIVSSAVCGTSTGEVTFTREPDVELPFA
Rv0127|M.tuberculosis_H37Rv --DDRTGFDALYDVAGPQFLLSLIVSSAVCGTSTGEVTFTREPDVELPFA
MMAR_0326|M.marinum_M --DDHTGFDALYDPVAPQVLLSLIDSSAVRSSSDGQVSFAREPDVELPLD
MUL_4796|M.ulcerans_Agy99 --DDHTGFDALYDPVAPQVLLSLIDSSAVRSSSDGQVSFAREPDVELPLD
MAV_5185|M.avium_104 --DDRTGFDALYDDEAPQFLLSLIDSSAVRSASGAEVRFAKEPDAQLPLE
MSMEG_6514|M.smegmatis_MC2_155 --DGQTAYDALYDPDATRYLLSLIDESATVQN----VRFVREPDVELPLD
TH_1781|M.thermoresistible__bu --GDRTAYDALYDPAAARHLLELFERSATVGA----VRFDKEPGVTLPLQ
Mflv_1087|M.gilvum_PYR-GCK PDGTRFAYDALFNPEAASRLLRLINSSETVGE----LTFSREPDVTLPVD
Mvan_5735|M.vanbaalenii_PYR-1 PQGERIAYDALFDPDAARHLLRLVDASATVAD----LRFTREPGATLPLY
. : .:***:: .. ** *. * . : * :**.. **.
Mb0132|M.bovis_AF2122/97 AQPRVCDAEQSNTSVIFDR------RAILKVFRRVSSGINPDIELNRVLT
Rv0127|M.tuberculosis_H37Rv AQPRVCDAEQSNTSVIFDR------RAILKVFRRVSSGINPDIELNRVLT
MMAR_0326|M.marinum_M AYPRVSDAEQSNTSVIFDRG----QAAILKVFRRVSSGINPDIELNRVLG
MUL_4796|M.ulcerans_Agy99 AYPRVSDAEQSNTSVIFDRG----QAAILKVFRRVSSGINPDIELNRVLG
MAV_5185|M.avium_104 AMAHVSDAEQSNTSVIFDR------DAIFKVFRRVSSGINPDIELNRVLG
MSMEG_6514|M.smegmatis_MC2_155 APPRVFGAEQSNTSVVFG------EDAIFKLFRRITPGVHPDIELNRVLA
TH_1781|M.thermoresistible__bu ASPRVSPAEQSNTSVIYEDVHGGEDAVIFKSFRRVTPGVHPDIELNRVLA
Mflv_1087|M.gilvum_PYR-GCK APAKVSGAEQSNTSVIFGK------DAMLKVFRRVTPGINPDIELNRVLA
Mvan_5735|M.vanbaalenii_PYR-1 APPKVSSAEQSNTSVIFGK------DAMLKVFRRVTPGINPDIELNRVLA
* .:* ********:: .::* ***::.*::*********
Mb0132|M.bovis_AF2122/97 RAGNPHVARLLGAYQFGRPNRSPTDALAYALGMVTEYEANAAEGWAMATA
Rv0127|M.tuberculosis_H37Rv RAGNPHVARLLGAYQFGRPNRSPTDALAYALGMVTEYEANAAEGWAMATA
MMAR_0326|M.marinum_M RAHNPHVARLLGTYEIGIPGEPP--EAACPLGMATAYAANAAEGWAMATA
MUL_4796|M.ulcerans_Agy99 RAHNPHVARLLGTYEIGIPGEPP--EAACPLGMATAYAANAAEGWAMATA
MAV_5185|M.avium_104 RAGNPHVARLLGTYEMAGADGTP--ETAWPLGMVTEFAANAAEGWAMATA
MSMEG_6514|M.smegmatis_MC2_155 RAGNPHVARLLGSFETEWE-GEP-----YALGMVTEFAANSAEGWDMATT
TH_1781|M.thermoresistible__bu RAGNPHVARLMAAYEIEFD-GAP-----YALGMITEFAANSAEGWDMATA
Mflv_1087|M.gilvum_PYR-GCK RAGNPRVATLLGSFETS----------SCALGMVTAFAANSAEGWDMATA
Mvan_5735|M.vanbaalenii_PYR-1 QAGNRHVARLLGSFETSWAGPGT---DRCALGMVTAFAANSAEGWDMATA
:* * :** *:.::: .*** * : **:**** ***:
Mb0132|M.bovis_AF2122/97 SVRDLFAEGDLYAHEVGGDFAGESYRLGEAVASVHATLADSLGTAQATFP
Rv0127|M.tuberculosis_H37Rv SVRDLFAEGDLYAHEVGGDFAGESYRLGEAVASVHATLADSLGTAQATFP
MMAR_0326|M.marinum_M SVRDLFAEGDLYAHEVGGDFAGESRRLGEAVASVHATLAEQLGTAQATFP
MUL_4796|M.ulcerans_Agy99 SVRDLFAEGDLYAHEVGGDFAGESRRLGEAVASVHATLAEQLGTAQATFP
MAV_5185|M.avium_104 SVRDLFAEGDLYAHEVGGDFAGESYRLGEAVASVHATLAETLGTSQAAFP
MSMEG_6514|M.smegmatis_MC2_155 STRDLFAEGDLYAEEVGGDFAGEAYRLGEAVASVHACLAHELGTEEVPFP
TH_1781|M.thermoresistible__bu STRDLYAEGDLYADEVGGDFAAESYRLGEAVASVHRSLAEQLGRGTAEFP
Mflv_1087|M.gilvum_PYR-GCK SVHDLFAN------EVGGDFADESRRLGEAVASVHATLAGALGTSVAEFP
Mvan_5735|M.vanbaalenii_PYR-1 SAREMFAD------VVGSDFADESYRLGNAVASVHATLAEALGTSTEPFP
*.::::*: **.*** *: ***:****** ** ** **
Mb0132|M.bovis_AF2122/97 VDRMLARLSSTVAVVPELREYAPTIEQQFQKLAAEAITVQRVHGDLHLGQ
Rv0127|M.tuberculosis_H37Rv VDRMLARLSSTVAVVPELREYAPTIEQQFQKLAAEAITVQRVHGDLHLGQ
MMAR_0326|M.marinum_M VDHVLARLSSTAAAVPELQQYAGTIEERFVKLVDETISVQRVHGDLHLGQ
MUL_4796|M.ulcerans_Agy99 VDHVLARLSSTAAAVPELQQYAGTIEERFVKLVDETISVQRVHGDLHLGQ
MAV_5185|M.avium_104 VDNVLARLSSTAALVPELTEYAATIEERFAKLATETITVQRVHGDLHLGQ
MSMEG_6514|M.smegmatis_MC2_155 ADVMAQRLAAAVDAVPELREHVPQIEERYHKLADTTMTVQRVHGDLHLGQ
TH_1781|M.thermoresistible__bu VEPLLARLAAATETVPALQPYAALIEERYRKLAEQTITVQRIHGDLHLGQ
Mflv_1087|M.gilvum_PYR-GCK IDTVLDRLRVATQAAPELAPYAPRIEERFRRLAERPIQVHRVHGDLHLGQ
Mvan_5735|M.vanbaalenii_PYR-1 VDTVLARLQSAARSAPELAGRAAAVEERYRRLDGRAITVQRVHGDLHLGQ
: : ** :. .* * . :*::: :* .: *:*:********
Mb0132|M.bovis_AF2122/97 VLRTPESWLLIDFEGEPGQPLDERRAPDSPLRDVAGVLRSFEYAAYGPLV
Rv0127|M.tuberculosis_H37Rv VLRTPESWLLIDFEGEPGQPLDERRAPDSPLRDVAGVLRSFEYAAYGPLV
MMAR_0326|M.marinum_M VLRTPESWVLIDFEGEPGQPLRERRAPDSPLRDVAGVLRSFEYAAYGPLV
MUL_4796|M.ulcerans_Agy99 VLRTPESWVLIDFEGESGQPLRERRAPDSPLRDVAGVLRSFEYAAYGPLV
MAV_5185|M.avium_104 VLRTPESWLLIDFEGEPGQPLEERRAPDSPLRDVAGVLRSFEYAAYGPLV
MSMEG_6514|M.smegmatis_MC2_155 VLRTPKGWLLIDFEGEPGQPLDERRRPDTPVRDVAGILRSFEYAAHQRLV
TH_1781|M.thermoresistible__bu VLRTPERWLLIDFEGEPGYPLSERRRPDSPLRDVAGMLRSYEYAAYHRLA
Mflv_1087|M.gilvum_PYR-GCK VLRTPESWLLIDFEGEPGQPLEDRRRPDSPLRDVAGVLRSFEYAAYQQVV
Mvan_5735|M.vanbaalenii_PYR-1 VLRTPDDWLLIDFEGEPGQPLDERRRPDSPLRDVAGVLRSFEYAAYQKLV
*****. *:*******.* ** :** **:*:*****:***:****: :.
Mb0132|M.bovis_AF2122/97 DQAT-----DKQLAARAREWVERNRAAFCDGYAVASGIDPRDSALLLGAY
Rv0127|M.tuberculosis_H37Rv DQAT-----DKQLAARAREWVERNRAAFCDGYAVASGIDPRDSALLLGAY
MMAR_0326|M.marinum_M DHAD-----DKQLAARAREWITRNRTAFCEGYAAASGNDPRDSELLLAAY
MUL_4796|M.ulcerans_Agy99 DHAD-----DKQLAARAREWITRNRTAFCEGYAAASGNDPRDSELLLAAY
MAV_5185|M.avium_104 EQGSQN--TDKQLAARAREWVERNRTAFCDGYAAASGIDPRDSAPLLAAY
MSMEG_6514|M.smegmatis_MC2_155 DQAGDDDDRARQLAARAREWVTRNCASFCDGYAAEAGTDPRDSADLLAAY
TH_1781|M.thermoresistible__bu EQGGDDLQHDKQLAARAREWVDRNTAAFCDGYAAVAGDDPRHHAELLAAY
Mflv_1087|M.gilvum_PYR-GCK TGG-----GDAQMAERARSWVDRNVDAFCAGYAAVAGEDPRASGDVLAAY
Mvan_5735|M.vanbaalenii_PYR-1 ELAPEQ-DADGRLADRARNWVDRNSAAFCAGYAAVAGDDPRRDGDVLAAY
. ::* ***.*: ** :** ***. :* *** :*.**
Mb0132|M.bovis_AF2122/97 ELDKAVYETGYETRHRPGWLPIPLRSIARLTAS
Rv0127|M.tuberculosis_H37Rv ELDKAVYETGYETRHRPGWLPIPLRSIARLTAS
MMAR_0326|M.marinum_M ELDKAVYEAGYESRHRPGWLPIPLRSIARLTAT
MUL_4796|M.ulcerans_Agy99 ELDKAVYEAGYESRHRPGWLPIPLRSIARLTAT
MAV_5185|M.avium_104 ELDKAVYEAGYEARHRPGWLPIPLRSIARLTAA
MSMEG_6514|M.smegmatis_MC2_155 ELDKAVYEAAYEARHRPSWLPIPLGSIARLLE-
TH_1781|M.thermoresistible__bu ELDKAVYEAGYEARHRPSWVPIPLESIARLIS-
Mflv_1087|M.gilvum_PYR-GCK ELDKAVYEAAYEARFRPSWLPIPMRSIQRLVS-
Mvan_5735|M.vanbaalenii_PYR-1 ELDKAVYEAAYEARFRPSWLPIPMRSIDRILG-
********:.**:*.**.*:***: ** *: