For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSPNAEVGRRAVLKVPVVVGAGLAISRTPRASAETARWSPERAHRWHRAQGWLVGANFIPANAINQLEMF QPGTFDPRRIDTELRMAKHLGLNTVRVFLHDLLWVQDRAGFQRRLARFVDIAAHHRIKPLFVLFDSCWDP HPRLGTQRGPVPGVHNSGWVQSPGAEHLGDPAYRRVLRDYVIGVISQFRHDKRVLGWDLWNEPDNPADVY REVERRDKVDRVAELLPQVFGWARSVDPVQPLTSGVWDGVWGDPARRTPVVRAQLDLSDVISFHSYADPR GFEDRLAELTPLGRPMLCTEYMARTLDSTVESILPIMKRRNIGAYTWGFVAGKTQTFLPWDSWERPVIEP PLWFHDLLHGDGTPYRAGEVNTIRELTGRNRPS
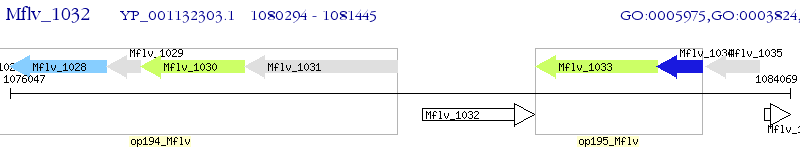
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1032 | - | - | 100% (383) | hypothetical protein Mflv_1032 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3123 | - | 1e-156 | 68.98% (374) | hypothetical protein Mb3123 |
| M. tuberculosis H37Rv | Rv3096 | - | 1e-156 | 68.98% (374) | hypothetical protein Rv3096 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1554 | - | 1e-158 | 67.29% (373) | hypothetical protein MMAR_1554 |
| M. avium 104 | MAV_0332 | - | 1e-152 | 67.91% (374) | hypothetical protein MAV_0332 |
| M. smegmatis MC2 155 | MSMEG_5877 | - | 1e-146 | 67.42% (356) | hypothetical protein MSMEG_5877 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_3476 | - | 1e-160 | 67.83% (373) | hypothetical protein MUL_3476 |
| M. vanbaalenii PYR-1 | Mvan_5801 | - | 0.0 | 83.55% (383) | hypothetical protein Mvan_5801 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3123|M.bovis_AF2122/97 ------MHRRTALKLPLLLAAGTVLGQAPRAAAGEPGRWSADRAHRWYQA
Rv3096|M.tuberculosis_H37Rv ------MHRRTALKLPLLLAAGTVLGQAPRAAAEEPGRWSADRAHRWYQA
MMAR_1554|M.marinum_M ------MQRRTALKLPLLLAAGAALAQPPRATA-VAGRWSAERANTWYQT
MUL_3476|M.ulcerans_Agy99 -----MVQRRTALKLPLLLAAGAALAQTPRATA-VAGRWSAERANTWYQT
MAV_0332|M.avium_104 ------MERRTALKLPLLLAAGAAVARAPRASAEEAGRWSPERANRWYQA
MSMEG_5877|M.smegmatis_MC2_155 ------------------------MTTAPRASA-APGQWPVERANAWYQA
Mflv_1032|M.gilvum_PYR-GCK MSPNAEVGRRAVLKVPVVVGAGLAISRTPRASA-ETARWSPERAHRWHRA
Mvan_5801|M.vanbaalenii_PYR-1 MPPEPLLNRRAALKVPAVLAAGMALSTVPRASA-ELTRWSPDRAHRWHRA
: ***:* :*. :**: *:::
Mb3123|M.bovis_AF2122/97 HGWLVGANYITSNAINQLEMFQPGTYDPRRIDNELGLARFHGFNTVRVFL
Rv3096|M.tuberculosis_H37Rv HGWLVGANYITSNAINQLEMFQPGTYDPRRIDNELGLARFHGFNTVRVFL
MMAR_1554|M.marinum_M QGWIVGANYITANAINQLEMFQPATYDPRRIDRELGLARLIGFNSMRVFL
MUL_3476|M.ulcerans_Agy99 QGWIVGANYITANAINQLEMFQPATYDPRRIDRELGLARLIGFNSMRVFL
MAV_0332|M.avium_104 QGWLVGANYIPANAINQLEMFQPDTFDPRRIDTELGWAQFYGHNTARVFL
MSMEG_5877|M.smegmatis_MC2_155 QGWLVGTNFITSNAINQLEMFSAGTYDPRRIDSELGACRLLGFNTVRVFL
Mflv_1032|M.gilvum_PYR-GCK QGWLVGANFIPANAINQLEMFQPGTFDPRRIDTELRMAKHLGLNTVRVFL
Mvan_5801|M.vanbaalenii_PYR-1 QGWLVGANFIPATAINQLEMFQPGTFDPRRIDSELRTAKLIGLNTVRVFL
:**:**:*:*.:.********.. *:****** ** .: * *: ****
Mb3123|M.bovis_AF2122/97 HDLLWAQDAPGFQTRLAQFVAIAARYHIKPLFVLFDSCWDPLPRPGRQRA
Rv3096|M.tuberculosis_H37Rv HDLLWAQDAPGFQTRLAQFVAIAARYHIKPLFVLFDSCWDPLPRPGRQRA
MMAR_1554|M.marinum_M HDQLWASDQRGFQTRLAQFVAIAARHGIKPLFVLFDSCWDPFPKLGQQRA
MUL_3476|M.ulcerans_Agy99 HDQLWASDQRGFQTRLAQFVAIAARHGIKPLFVLFDSCWDPFPKLGQQRA
MAV_0332|M.avium_104 HDQLWAADQRGFQTRLGQFVDIAARHRIKPLFVFFDSCWDPQPRAGRQRP
MSMEG_5877|M.smegmatis_MC2_155 HDLLWAQDRAGFQNRLAQFVSIASRQGIKPLFVLFDSCWDPLPKPGAQRA
Mflv_1032|M.gilvum_PYR-GCK HDLLWVQDRAGFQRRLARFVDIAAHHRIKPLFVLFDSCWDPHPRLGTQRG
Mvan_5801|M.vanbaalenii_PYR-1 HDLLWVQDRVGFQRRLARFVDIAAHHGIKPLFVLFDSCWDPHPRLGKQRD
** **. * *** **.:** **:: ******:******* *: * **
Mb3123|M.bovis_AF2122/97 PRAGVHNSGWVQSPGAERLDDRRYASTLYNYVTGVLGQFRNDDRVLGWDL
Rv3096|M.tuberculosis_H37Rv PRAGVHNSGWVQSPGAERLDDRRYASTLYNYVTGVLGQFRNDDRVLGWDL
MMAR_1554|M.marinum_M PRPGVHNSGWVQSPGAEHLGDPSYQAVLHGYVTGVLNQFRNDNRVLGWDL
MUL_3476|M.ulcerans_Agy99 PTPGVHNSGWVQSPGAEHLGDPSYQAVLHGYVTGVLNQFRNDNRVLGWDL
MAV_0332|M.avium_104 PRPGVHNSGWVQSPGAERLGDPRYIPVMRDYVTSVMTQFRNDDRVLGWDL
MSMEG_5877|M.smegmatis_MC2_155 PTPGVHNSGWVQSPGAQRIDDPRYRPVLRDYVVGVMSQFRNDQRVLGWDL
Mflv_1032|M.gilvum_PYR-GCK PVPGVHNSGWVQSPGAEHLGDPAYRRVLRDYVIGVISQFRHDKRVLGWDL
Mvan_5801|M.vanbaalenii_PYR-1 PIPGVHNSGWVQSPGAEHLSDPRHRRVLRDYVVGVLSQFRHDKRVLGWDL
* .*************:::.* : .: .** .*: ***:*.*******
Mb3123|M.bovis_AF2122/97 WNEPDNPARVYRKVERKDKLERVAELLPQVFRWARTVDPVQPLTSGVWQG
Rv3096|M.tuberculosis_H37Rv WNEPDNPARVYRKVERKDKLERVAELLPQVFRWARTVDPVQPLTSGVWQG
MMAR_1554|M.marinum_M WNEPDNPAKVYRKVERKDKLERVAELLPQVFQWAREVDPSQPLTSGVWQG
MUL_3476|M.ulcerans_Agy99 WNEPDNPAKVYRKVERKDKLERVAELLPQVFQWAREVDPSQPLTSGVWQG
MAV_0332|M.avium_104 WNEPDNPARQYRNVERSDKEQLVANLLPQVFRWARAVDASQPLTSGVWRG
MSMEG_5877|M.smegmatis_MC2_155 WNEPDNPARQYRKVERSDKLDAVGALLPQVFGWARSVNAAQPLTSGVWQG
Mflv_1032|M.gilvum_PYR-GCK WNEPDNPADVYREVERRDKVDRVAELLPQVFGWARSVDPVQPLTSGVWDG
Mvan_5801|M.vanbaalenii_PYR-1 WNEPDNPADAYKDVERRDKVDRVAELLPQVFQWARSVDPVQPLTSGVWDG
******** *:.*** ** : *. ****** *** *:. ******** *
Mb3123|M.bovis_AF2122/97 NWGDPGRRSTISAIQLDNADVITFHSYAAPAEFEGRIAELAPLQRPILCT
Rv3096|M.tuberculosis_H37Rv NWGDPGRRSTISAIQLDNADVITFHSYAAPAEFEGRIAELAPLQRPILCT
MMAR_1554|M.marinum_M NWSDPGKRSTIASIQLDNADVITFHSYAAPAGFEARIDELAPLGRPIICT
MUL_3476|M.ulcerans_Agy99 NWSDPGKRSTIASIQLDNADVITFHSYAAPAGFEARIDELAPLGRPIICT
MAV_0332|M.avium_104 DWGQPQGRSAISDIQLANADVITFHSYAEPAGFESRINELTPLGRPILCT
MSMEG_5877|M.smegmatis_MC2_155 SWER-GRRSEMASFQLDNSDVISFHSYAGPDEFEARIAELEPLGRPILCT
Mflv_1032|M.gilvum_PYR-GCK VWGDPARRTPVVRAQLDLSDVISFHSYADPRGFEDRLAELTPLGRPMLCT
Mvan_5801|M.vanbaalenii_PYR-1 EWGDPARRNEINRIQLDLSDVITFHSYADRRGFEARLEELTPIGRPMLCT
* *. : ** :***:***** ** *: ** *: **::**
Mb3123|M.bovis_AF2122/97 EYLARSQGSTVEGILPIAKRHNVGAFNWGLVAGKTQTYLPWDSWDHPYRA
Rv3096|M.tuberculosis_H37Rv EYLARSQGSTVEGILPIAKRHNVGAFNWGLVAGKTQTYLPWDSWDHPYRA
MMAR_1554|M.marinum_M EYLARSQGSSVEGVLPIAKRRNVGAFNWGLVAGKTQTYLPWDSWDHPYTK
MUL_3476|M.ulcerans_Agy99 EYLARSQDSSVEGVLPIAKRRNVGAFNWGLVAGKTQTYLPWDSWDHPYTK
MAV_0332|M.avium_104 EYMARPRGSTVESILPVAKRHNVGAINWGLVAGKTQTYFPWESWDHPYTS
MSMEG_5877|M.smegmatis_MC2_155 EYLARSEGSTLEGVLPVAKRHNVGAYSWGLVAGKTQTYFPWDSWDKPYTK
Mflv_1032|M.gilvum_PYR-GCK EYMARTLDSTVESILPIMKRRNIGAYTWGFVAGKTQTFLPWDSWERPVIE
Mvan_5801|M.vanbaalenii_PYR-1 EYMARTLDSTVETILPITRRRNVGAYTWGFFAGKTQTFLPWDSWDRPVTG
**:**. .*::* :**: :*:*:** .**:.******::**:**::*
Mb3123|M.bovis_AF2122/97 PPKVWFHDLLHPNGRPYRDGEVQTIRKLNGMPSQD
Rv3096|M.tuberculosis_H37Rv PPKVWFHDLLHPNGRPYRDGEVQTIRKLNGMPSQD
MMAR_1554|M.marinum_M PPKVWFSDLLQPDGRPYRESEIQTIQSLTGARTQD
MUL_3476|M.ulcerans_Agy99 PPKVWFSDLLQPDGRPYRESEIQTIQSLTGARTQD
MAV_0332|M.avium_104 VPKVWFHDLIRPEGRPFQDIEALTVRKLAGSPT--
MSMEG_5877|M.smegmatis_MC2_155 VPNVWFHDLLRPDGRPYKDSEYATLRKLTARV---
Mflv_1032|M.gilvum_PYR-GCK PP-LWFHDLLHGDGTPYRAGEVNTIRELTGRNRPS
Mvan_5801|M.vanbaalenii_PYR-1 PPGLWFHDLLNGDGSPYRDSEINTIRELTGRRGP-
* :** **:. :* *:: * *::.* .