For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDGAVTAEYLRNFVFDKPSFGKRGYNEKAVADFVALCARRLDGRGHLTAADVRNVRFNKPPLGRRGYDDA QVDQLLTEIATAIDKLDA*
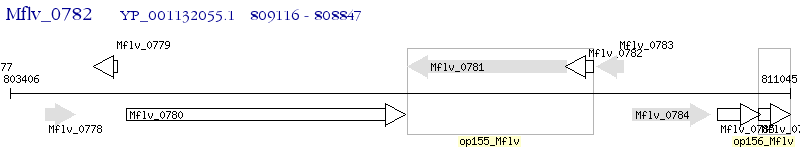
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0782 | - | - | 100% (89) | hypothetical protein Mflv_0782 |
| M. gilvum PYR-GCK | Mflv_2996 | - | 9e-07 | 47.50% (40) | DivIVA family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2169c | wag31 | 9e-07 | 47.50% (40) | hypothetical protein Mb2169c |
| M. tuberculosis H37Rv | Rv2145c | wag31 | 9e-07 | 47.50% (40) | hypothetical protein Rv2145c |
| M. leprae Br4923 | MLBr_00922 | ag84 | 5e-07 | 47.50% (40) | immunogenic protein, antigen 84 |
| M. abscessus ATCC 19977 | MAB_2017 | - | 1e-06 | 47.50% (40) | immunogenic protein antigen 84 |
| M. marinum M | MMAR_3185 | wag31 | 1e-06 | 47.50% (40) | secreted antigen Wag31 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0050 | - | 2e-20 | 50.62% (81) | hypothetical protein MSMEG_0050 |
| M. thermoresistible (build 8) | TH_3558 | - | 1e-19 | 53.01% (83) | PUTATIVE conserved hypothetical protein |
| M. ulcerans Agy99 | MUL_3493 | wag31 | 9e-07 | 47.50% (40) | secreted antigen Wag31 |
| M. vanbaalenii PYR-1 | Mvan_0065 | - | 2e-40 | 85.39% (89) | hypothetical protein Mvan_0065 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3185|M.marinum_M -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELTRLIEE
MUL_3493|M.ulcerans_Agy99 -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELTRLIEE
Mb2169c|M.bovis_AF2122/97 -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELTRLIEE
Rv2145c|M.tuberculosis_H37Rv -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELTRLIEE
MLBr_00922|M.leprae_Br4923 -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELTQLIEE
MAB_2017|M.abscessus_ATCC_1997 -----MPLTPADVHNVAFSKPPIGKRGYNEDEVDAFLDLVENELARLIEE
Mflv_0782|M.gilvum_PYR-GCK MSDG--AVTAEYLRNFVFDKPSFGKRGYNEKAVADFVALCARRLD-----
Mvan_0065|M.vanbaalenii_PYR-1 MSDG--AVTAEYLRNFVFEKPPWGKRGYNEKAVGDFVALCARRLD-----
TH_3558|M.thermoresistible__bu MGEVTAGLTAADVRNTRFEKPGWGRRGYHPKAVDDLLMLVARRLD-----
MSMEG_0050|M.smegmatis_MC2_155 MSAMSRVVSADDVRTVEFAKPPIGKRGYDKKSVDDFLQLVARRLE-----
::. ::. * ** *:***. . * :: * ..*
MMAR_3185|M.marinum_M NSDLRQRIAELDQELAAGA--GGGAAVTAQPTQAMPVYEPEPEPAKPAAP
MUL_3493|M.ulcerans_Agy99 NSDLRQRIAELDQELAAGA--GGGAAVTAQPTQAMPVYEPEPEPAKPAAP
Mb2169c|M.bovis_AF2122/97 NSDLRQRINELDQELAAG----GGAGVTPQATQAIPAYEPE--PGKPAPA
Rv2145c|M.tuberculosis_H37Rv NSDLRQRINELDQELAAG----GGAGVTPQATQAIPAYEPE--PGKPAPA
MLBr_00922|M.leprae_Br4923 NSDLRQRIEELDHELAAGGGTGAGPVIAVQPTQALSTFEPELVSAKQAPV
MAB_2017|M.abscessus_ATCC_1997 NSDLRQRVQELDQELAEAR-KGGGAAPVYEAPKPEKKPEPAKPEPTPAPV
Mflv_0782|M.gilvum_PYR-GCK ----------------------GRGHLTAADVRNVR---------FNKPP
Mvan_0065|M.vanbaalenii_PYR-1 ----------------------GRGHLSAEDVRNVR---------FNKPP
TH_3558|M.thermoresistible__bu ----------------------GRGYLCADDLRGLR---------FPKAP
MSMEG_0050|M.smegmatis_MC2_155 ----------------------GRGHLSADDVRNIA---------FPKPP
. : .
MMAR_3185|M.marinum_M VASA---ATNEEQAMKAARVLSLAQDTADRLTSTAKAESDKMLSDARANA
MUL_3493|M.ulcerans_Agy99 VASA---ATNEEQAMKAARVLSLAQDTADRLTSTAKAESDKMLSDARANA
Mb2169c|M.bovis_AF2122/97 AVSA---GMNEEQALKAARVLSLAQDTADRLTNTAKAESDKMLADARANA
Rv2145c|M.tuberculosis_H37Rv AVSA---GMNEEQALKAARVLSLAQDTADRLTNTAKAESDKMLADARANA
MLBr_00922|M.leprae_Br4923 AAVA---ETAEELAMKATRVLSLAQDTADQLTSTAKVESDKMLADARVNA
MAB_2017|M.abscessus_ATCC_1997 VAAAPAGASTEEHHVRAAKILGLAQDTADRLTGNAKSESEKMLADARANA
Mflv_0782|M.gilvum_PYR-GCK LGRR-----------------GYDDAQVDQLLTEIATAIDKLDA------
Mvan_0065|M.vanbaalenii_PYR-1 FGKR-----------------GFDEAQVDALLGEIATAIAKLDA------
TH_3558|M.thermoresistible__bu LLRR-----------------GYDRAAVDEFLERMTAAVAALETNG----
MSMEG_0050|M.smegmatis_MC2_155 VFQR-----------------GYSEDEVDALLDDVVATLER---------
. .* :
MMAR_3185|M.marinum_M DQILSEARHTAETTVTEARQRADGMLADAQARSESQLRQAQEKADALQAD
MUL_3493|M.ulcerans_Agy99 DQILSEARHTAETTVTEARQRADGMLADAQARSESQLRQAQEKADALQAD
Mb2169c|M.bovis_AF2122/97 EQILGEARHTADATVAEARQRADAMLADAQSRSEAQLRQAQEKADALQAD
Rv2145c|M.tuberculosis_H37Rv EQILGEARHTADATVAEARQRADAMLADAQSRSEAQLRQAQEKADALQAD
MLBr_00922|M.leprae_Br4923 DQILGEARLTAEATVAEAQQRADAMLADAQTRSEVQSRQAQEKADALQAE
MAB_2017|M.abscessus_ATCC_1997 DQIVSEARATAEKTVTEARTKAEALLSDAQTRSETQLRQAQEKADALQSD
Mflv_0782|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0065|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3558|M.thermoresistible__bu --------------------------------------------------
MSMEG_0050|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3185|M.marinum_M AERKHSEIMGTINQQRTVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
MUL_3493|M.ulcerans_Agy99 AERKHSEIMGTINQQRTVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
Mb2169c|M.bovis_AF2122/97 AERKHSEIMGTINQQRAVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
Rv2145c|M.tuberculosis_H37Rv AERKHSEIMGTINQQRAVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
MLBr_00922|M.leprae_Br4923 AERKHSEIMGAISQQRTVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
MAB_2017|M.abscessus_ATCC_1997 AERKHSEIMGTINQQRTVLEGRLEQLRTFEREYRTRLKTYLESQLEELGQ
Mflv_0782|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0065|M.vanbaalenii_PYR-1 --------------------------------------------------
TH_3558|M.thermoresistible__bu --------------------------------------------------
MSMEG_0050|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3185|M.marinum_M RGSAAPVDSSADAG------GFEQFNRGNN-
MUL_3493|M.ulcerans_Agy99 RGSAAPVDSSADAG------GFEQFNRGNN-
Mb2169c|M.bovis_AF2122/97 RGSAAPVDSNADAG------GFDQFNRGKN-
Rv2145c|M.tuberculosis_H37Rv RGSAAPVDSNADAG------GFDQFNRGKN-
MLBr_00922|M.leprae_Br4923 RGSAAPVDSNADAG------GFDQFNRGNN-
MAB_2017|M.abscessus_ATCC_1997 RGSAAPVDAGASNENASKEAGFGQFNRGNNY
Mflv_0782|M.gilvum_PYR-GCK -------------------------------
Mvan_0065|M.vanbaalenii_PYR-1 -------------------------------
TH_3558|M.thermoresistible__bu -------------------------------
MSMEG_0050|M.smegmatis_MC2_155 -------------------------------