For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QPLNHNPAAAGIGGQVTANGARGLGVGTAATAEVSALAPAGVDEISAVAALSFASEGIQTLGINAMAQQE IARAGATVIEASVAYEATDAANGAKLI*
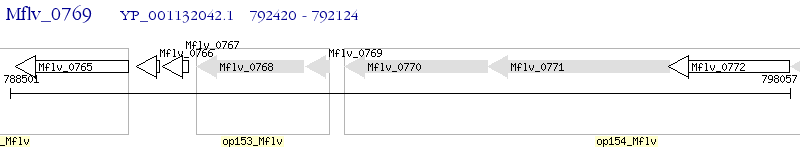
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0769 | - | - | 100% (98) | PE-like protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3902 | PE35 | 1e-17 | 45.56% (90) | PE family-like protein |
| M. tuberculosis H37Rv | Rv3872 | PE35 | 1e-17 | 45.56% (90) | PE family-related protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2231c | - | 1e-06 | 33.78% (74) | PE family protein |
| M. marinum M | MMAR_0185 | - | 9e-19 | 46.67% (90) | PE family protein, PE35 |
| M. avium 104 | MAV_0117 | - | 6e-05 | 32.94% (85) | PE family protein |
| M. smegmatis MC2 155 | MSMEG_0063 | - | 2e-26 | 55.67% (97) | PE family protein |
| M. thermoresistible (build 8) | TH_3343 | - | 3e-25 | 60.47% (86) | PUTATIVE PE domain protein |
| M. ulcerans Agy99 | MUL_4359 | - | 2e-14 | 38.78% (98) | PE family protein |
| M. vanbaalenii PYR-1 | Mvan_0076 | - | 2e-37 | 73.47% (98) | PE-like protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3902|M.bovis_AF2122/97 MEKMSHDPIAADIGTQVSDNALHGVTAGSTALTSVTGLVPAGADEVSAQA
Rv3872|M.tuberculosis_H37Rv MEKMSHDPIAADIGTQVSDNALHGVTAGSTALTSVTGLVPAGADEVSAQA
MUL_4359|M.ulcerans_Agy99 MQSMSIDPVAADIGAQLAEGAFRGLQAGATAATSITSVRPAGADEVSTQA
MMAR_0185|M.marinum_M MRSMSFDPAAASVAAAISAHASRGLDAGTAAASSVTGLAPAGADEISAQF
Mflv_0769|M.gilvum_PYR-GCK MQPLNHNPAAAGIGGQVTANGARGLGVGTAATAEVSALAPAGVDEISAVA
Mvan_0076|M.vanbaalenii_PYR-1 MQPLSHNPGAIGVGGQVTANGARGLATGTAATSAVSALAPAGADEVSAAA
MSMEG_0063|M.smegmatis_MC2_155 MQPMTHNPGAEAVAAQVIANAARGLAGGTTASAAVTALVPAGADEVSALA
TH_3343|M.thermoresistible__bu ----------VGIGSQVIANGTRGLAVGTAASAEVTALVPAGADEVSAQA
MAB_2231c|M.abscessus_ATCC_199 MNLNVVPEGLTATSAAVEALTARLAAAHAAAAPVIGAVVPPAADPVSLET
MAV_0117|M.avium_104 MTLRVIPEGLAATSAAVDAITAQLAAVHAAAAPVVSVVVPPAADPVSLAA
. : : ::* . : : *...* :*
Mb3902|M.bovis_AF2122/97 ATAFTSEGIQLLASNASAQDQLHRAGEAVQDVARTYSQIDDGAAG-----
Rv3872|M.tuberculosis_H37Rv ATAFTSEGIQLLASNASAQDQLHRAGEAVQDVARTYSQIDDGAAG-----
MUL_4359|M.ulcerans_Agy99 MLAFTKHAGQMLALNQAAQEELRRAGEAVNAIARMYADTDVAVARNLIDV
MMAR_0185|M.marinum_M AAAFAAEGAQVLALNTAAQDELARAGQALRQIAGMYSAVDNSWSD-----
Mflv_0769|M.gilvum_PYR-GCK ALSFASEGIQTLGINAMAQQEIARAGATVIEASVAYEATDAANGA-----
Mvan_0076|M.vanbaalenii_PYR-1 AVTFAAEGIQTLGINALAQEEIARAGASIIEAAGAYQAVDASNAT-----
MSMEG_0063|M.smegmatis_MC2_155 AVAFASEGVEALAANAFAQEELTRAGAAFAEIAGIYNAVDAANAA-----
TH_3343|M.thermoresistible__bu AAAFAKEGMEALALNTFAQEELARAGAAVVQIAGIYDAVDAANAG-----
MAB_2231c|M.abscessus_ATCC_199 AAGFSARGIEHSGVAAQAVEELGRAGLGVSESAASYTTGDMQAAA-----
MAV_0117|M.avium_104 AAVFSERGSQHSAAATKGVEVLGRAGLGVSLAGVNYVAGDAASAS-----
*: .. : . . : : *** . . * * .
Mb3902|M.bovis_AF2122/97 ------VFA----
Rv3872|M.tuberculosis_H37Rv ------VFAE---
MUL_4359|M.ulcerans_Agy99 GWRSGSALANV--
MMAR_0185|M.marinum_M ------TLA----
Mflv_0769|M.gilvum_PYR-GCK ------KLI----
Mvan_0076|M.vanbaalenii_PYR-1 ------TLI----
MSMEG_0063|M.smegmatis_MC2_155 ------TM-----
TH_3343|M.thermoresistible__bu ------TLA----
MAB_2231c|M.abscessus_ATCC_199 ------AYMIARG
MAV_0117|M.avium_104 ------TYLSAG-