For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
IRTAARAVDAERIKLLTLRSPLWAAAAAAVLSFALAALQASVSYGYAAMPVSTAAMGVAVFGVPVLMVVA AMTVTGEYRSGLIATTFTAIPRRTLVVCAKAVVAAAFSVVASAVMVFGSVMVARAAAAPGVAAGLTVSAT MRFAAAIAVYAALTAVLGVGVAVLLRHTAGAVTVLLLWPLLIEPLLGNLPGRSVEIGPYLPFANVFRFLD VQWLFPEYPMHWGTLGSLGYFAAVVGVVFAAGLVMVNRRDA*
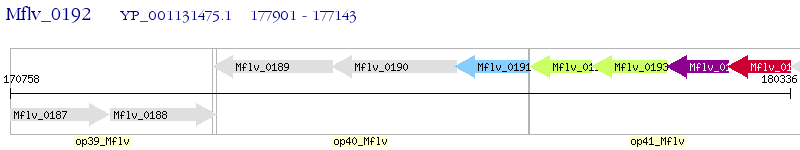
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0192 | - | - | 100% (252) | putative ABC transporter membrane protein |
| M. gilvum PYR-GCK | Mflv_4300 | - | 9e-07 | 23.83% (193) | hypothetical protein Mflv_4300 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4214c | - | 1e-49 | 45.93% (246) | ABC transporter |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_0796 | - | 5e-81 | 60.56% (251) | ABC transporter integral membrane subunit |
| M. thermoresistible (build 8) | TH_3177 | - | 3e-73 | 55.82% (249) | PUTATIVE putative ABC transporter integral membrane subunit |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0713 | - | 1e-100 | 74.50% (251) | putative ABC transporter membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0192|M.gilvum_PYR-GCK ------MIRTAARAVDAERIKLLTLRSPLWAAAAAAVLSFALAALQASVS
Mvan_0713|M.vanbaalenii_PYR-1 ------MMATAGAVLNAERIKLTTLRSPLWATVAAAVLSFGLSALQASVA
MSMEG_0796|M.smegmatis_MC2_155 --------MSALAALDAERIKLSTTRSPLWSVVGAAVLSFAIAALQGWSA
TH_3177|M.thermoresistible__bu VSAVMSMATGLAAAVNAERVKLLTTRAPLWSALGVAVSALGVAALQGLTA
MAB_4214c|M.abscessus_ATCC_199 -----------MGVVAAERIKVCTSRSAVWCSLLALALGVGLAGLQGGSA
.: ***:*: * *:.:*. . . ...::.**. :
Mflv_0192|M.gilvum_PYR-GCK YGYAAMPVSTAAMGVAVFGVPVLMVVAAMTVTGEYRSGLIATTFTAIPRR
Mvan_0713|M.vanbaalenii_PYR-1 YDYARLTVPDAALGVAVFGVPVLMVVSAMTVTGEYRSGLIATTFMATPHR
MSMEG_0796|M.smegmatis_MC2_155 YGYTPLPPGKAALGVAVFGVPVLMVLASMTMTGEYRTGLIRTTFMATPNR
TH_3177|M.thermoresistible__bu HHYSALPPQQAALGVVVFGIPVLMILAALTVTGEHRTGAIRTTFLAVPRR
MAB_4214c|M.abscessus_ATCC_199 SIYAPISPGDAAGGAALVGVPLLMVLAAMTVTNENRTAMVRTTFQATPNR
*: :. ** *..:.*:*:**:::::*:*.* *:. : *** * *.*
Mflv_0192|M.gilvum_PYR-GCK TLVVCAKAVVAAAFSVVASAVMVFGSVMVARAAAAPGVAAGLTVS--ATM
Mvan_0713|M.vanbaalenii_PYR-1 TLVFCAKALVAAVFSALSAAVMVVGSVLVARLVADPGVGDALTPA--ATA
MSMEG_0796|M.smegmatis_MC2_155 SVVLAAKAIVCAAFSSVAAIVMVLGAVLVARLFTDPLIAVDLSPTNPATW
TH_3177|M.thermoresistible__bu TVAIAAKTVVATVFCGGWAALMTVAAILAAGASADPMAAAHLTPTGAAGW
MAB_4214c|M.abscessus_ATCC_199 LTVIGAKALVAGGWVAVVTAVTALTSMALVRAMAGPVAGANLALDSTGVW
.. **::*. : : : .. :: . : * . *: .
Mflv_0192|M.gilvum_PYR-GCK RFAAAIAVYAALTAVLGVGVAVLLRHTAGAVTVLLLWPLLIEPLLGNLPG
Mvan_0713|M.vanbaalenii_PYR-1 GFVAAIALYAAPAAVLGVGVATLLRHTAGAVTVLLLWPLLIEPLLANLPG
MSMEG_0796|M.smegmatis_MC2_155 QVAGSFALYAALAAILGVAVGALVRFSAGAVAVLLLWPLVAEPLLANMPG
TH_3177|M.thermoresistible__bu RVIAGVTAFAMLAAVLGVAVGVLLRNTAGAVAVLLLWPLVAEPILANMPD
MAB_4214c|M.abscessus_ATCC_199 NTVGAVTVYAFLCAVLAVAVGVLLKHTAGAVAVLLLWPTVLELMLGWLTD
...: :* *:*.*.*..*:: :****:****** : * :*. :..
Mflv_0192|M.gilvum_PYR-GCK RSVEIGPYLPFANVFRFLDVQWLFPE-YPMHWGTLGSLGYFAAVVGVVFA
Mvan_0713|M.vanbaalenii_PYR-1 RGWQIGPYLPFANMFRFLDVQWLFPE-FAMHWGGPGSLGYFTAVVATVFV
MSMEG_0796|M.smegmatis_MC2_155 NGAQIGPWLPFVHMYRFLDVDWLFPS-YAMPWGPVGSLIYFVVLVVVVFV
TH_3177|M.thermoresistible__bu VAPVVGPYLPFGNVFRFLDVRWLYPV-FDTPWGGLGSLVYFAVVVAVVGV
MAB_4214c|M.abscessus_ATCC_199 AGKALQPFLPFANGIRFTGVPWYTSDGVRFVWDATGSLIYFATVVVITLV
. : *:*** : ** .* * . *. *** **..:* . .
Mflv_0192|M.gilvum_PYR-GCK AGLVMVNRRDA
Mvan_0713|M.vanbaalenii_PYR-1 AALITVNRRDT
MSMEG_0796|M.smegmatis_MC2_155 AAVVALNRRDA
TH_3177|M.thermoresistible__bu AAVVVVNRRDA
MAB_4214c|M.abscessus_ATCC_199 AALAVVQRRDV
*.: ::***.