For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEQQWNFAGIEAAASAIQGNVTSIHSLLDEGKQSLTKLAAAWGGSGSEAYQGVQQKWDATATELNNALQ NLARTISEAGQAMASTEGNVTGMFA
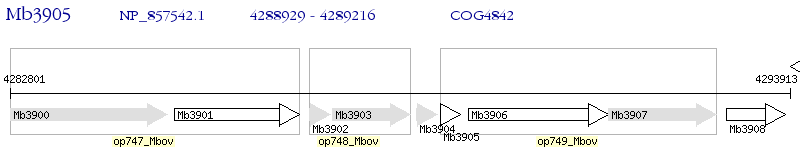
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3905 | esxA | - | 100% (95) | 6 kDa early secretory antigenic target EsaT6 (EsaT-6) |
| M. bovis AF2122 / 97 | Mb3045c | esxR | 2e-05 | 26.14% (88) | putative secreted ESAT-6 like protein 9 TB10.3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0766 | - | 3e-19 | 47.78% (90) | early secretory antigenic target, 6 kDa |
| M. tuberculosis H37Rv | Rv3875 | esxA | 4e-50 | 100.00% (95) | 6 kDa early secretory antigenic target ESXA (ESAT-6) |
| M. leprae Br4923 | MLBr_00049 | - | 1e-17 | 38.46% (91) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_2326c | - | 1e-10 | 34.44% (90) | hypothetical protein MAB_2326c |
| M. marinum M | MMAR_5450 | esxA | 5e-46 | 91.58% (95) | 6 kDa early secretory antigenic target EsxA (EsaT-6) |
| M. avium 104 | MAV_4387 | - | 1e-06 | 27.47% (91) | hypothetical protein MAV_4387 |
| M. smegmatis MC2 155 | MSMEG_0066 | - | 4e-34 | 71.58% (95) | early secretory antigenic target, 6 kDa |
| M. thermoresistible (build 8) | TH_2268 | esat6 | 2e-30 | 65.26% (95) | 6 KDA EARLY SECRETORY ANTIGENIC TARGET ESXA (ESAT-6) |
| M. ulcerans Agy99 | MUL_2243 | esxR | 8e-06 | 27.59% (87) | EsaT-6 family protein |
| M. vanbaalenii PYR-1 | Mvan_0079 | - | 1e-16 | 43.82% (89) | 6 kDa early secretory antigenic target EsaT6 (EsaT-6) |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3905|M.bovis_AF2122/97 -MTEQQWNFAGIEAAAS-AIQGNVTSIHSLLDEGKQSLTKLAAAWGGSGS
Rv3875|M.tuberculosis_H37Rv -MTEQQWNFAGIEAAAS-AIQGNVTSIHSLLDEGKQSLTKLAAAWGGSGS
MMAR_5450|M.marinum_M -MTEQQWNFAGIEAASS-AIQGNVTSIHSLLDEGKQSLHKLAAAWGGSGS
MSMEG_0066|M.smegmatis_MC2_155 -MTEQVWNFAGIEGGAS-EIHGAVSTTAGLLDEGKASLTTLASAWGGTGS
TH_2268|M.thermoresistible__bu -MSQQVWNFAGIEGGAG-EIHAAVGTTAGLLDEGKASLAALASAWGGSGS
MLBr_00049|M.leprae_Br4923 --MIQAWHFPALQGAVN-ELQGSQSRIDALLEQCQESLTKLQSSWHGSGN
Mflv_0766|M.gilvum_PYR-GCK --MALAYNFAGIDNNAG-EIMGAVGRTEGLLQEGQGSLARLSAVWGGTAS
Mvan_0079|M.vanbaalenii_PYR-1 ---MITYNFAGIESDAG-EISGQVGKVAGLLAEGHGALTRLQAVWTGDGQ
MAB_2326c|M.abscessus_ATCC_199 -MADLVYDYAKISALVG-DWQASIARLQQLLDEQRAEVNKLAESWTGATG
MUL_2243|M.ulcerans_Agy99 -MSQIMYNYPAMLAHAG-DMSGYAGSLHGLGADIAAEQAALSNAWQGDTG
MAV_4387|M.avium_104 MDPTLSYNFGEIEHSVRQEIHTTSARFNAALDELRARIAPLQQLWTSEAA
:.: : : * * .
Mb3905|M.bovis_AF2122/97 EAYQGVQQKWDATATELNNALQNLARTISEAGQAMASTEGNVTG--MFA-
Rv3875|M.tuberculosis_H37Rv EAYQGVQQKWDATATELNNALQNLARTISEAGQAMASTEGNVTG--MFA-
MMAR_5450|M.marinum_M EAYRGVQQNWDSTAQELNNSLQNLARTISEAGQAMSSTEGNVTG--MFA-
MSMEG_0066|M.smegmatis_MC2_155 EAYQAVQARWDSTSNELNLALQNLAQTISEAGQTMAQTEAGVTG--MFA-
TH_2268|M.thermoresistible__bu EAYQAVQMRWDSVSAELNAALQNLAQTISEAGANMAQTEAGVTG--MFA-
MLBr_00049|M.leprae_Br4923 ESYSSVQQRFNQNTEGINHALGDLVQAINHSAETMQQTEAGVTS--MFTG
Mflv_0766|M.gilvum_PYR-GCK DAYVAVQNRWDSSSNELNMALKSLANAIAQAGHDMGATEMRNQG--KFA-
Mvan_0079|M.vanbaalenii_PYR-1 MAYDAVQNRWNQNSEELNLALQSLSEALSRSGGDMFGADQGVAG--QFT-
MAB_2326c|M.abscessus_ATCC_199 DAFRREQKIWQQTADTLLAAAKKLGGLISAAAEAMAKTEAGVANNPQFSG
MUL_2243|M.ulcerans_Agy99 VTYQGWQTQWNQAMEDLVRAYQSMASTHESNTLAMSARDQAEAA--KWGA
MAV_4387|M.avium_104 TAYQAEQLKWHRSATALNEILVQLGDAVRDGAEEVADADRRAAG--VWAR
:: * :. : .: : : :
Mb3905|M.bovis_AF2122/97 --
Rv3875|M.tuberculosis_H37Rv --
MMAR_5450|M.marinum_M --
MSMEG_0066|M.smegmatis_MC2_155 --
TH_2268|M.thermoresistible__bu --
MLBr_00049|M.leprae_Br4923 --
Mflv_0766|M.gilvum_PYR-GCK --
Mvan_0079|M.vanbaalenii_PYR-1 --
MAB_2326c|M.abscessus_ATCC_199 SD
MUL_2243|M.ulcerans_Agy99 --
MAV_4387|M.avium_104 --