For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPLFSFEGRSPRIDPTAFVAPTATLIGDVTIEAGASVWFNAVLRGDYAPVVVREGANVQDGAVLHAPPGI PVDIGPGATVAHLCVIHGVHVGSEALIANHATVLDGAVIGARCMIAAGALVVAGTQIPAGMLVTGAPAKV KGPIEGTGAEMWVNVNPQAYRDLAARHLAGLEPM
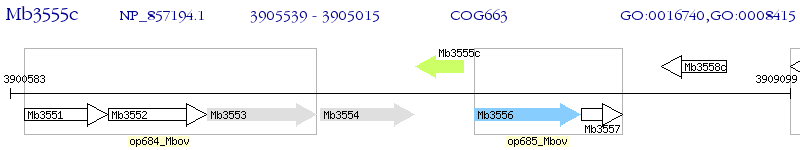
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3555c | - | - | 100% (174) | siderophore-binding protein |
| M. bovis AF2122 / 97 | Mb3292c | manC | 6e-06 | 30.70% (114) | mannose-1-phosphate guanyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2121 | - | 7e-34 | 41.21% (165) | carbonic anhydrase |
| M. tuberculosis H37Rv | Rv3525c | - | 2e-98 | 100.00% (174) | siderophore-binding protein |
| M. leprae Br4923 | MLBr_00753 | rmlA2 | 4e-08 | 34.21% (114) | putative sugar-phosphate nucleotidyl transferase |
| M. abscessus ATCC 19977 | MAB_4172c | - | 6e-65 | 65.12% (172) | siderophore-binding protein |
| M. marinum M | MMAR_5014 | - | 6e-93 | 91.38% (174) | hypothetical protein MMAR_5014 |
| M. avium 104 | MAV_0634 | - | 8e-85 | 83.33% (174) | carnitine operon protein CaiE |
| M. smegmatis MC2 155 | MSMEG_5191 | - | 1e-33 | 43.75% (160) | siderophore binding protein |
| M. thermoresistible (build 8) | TH_1984 | - | 2e-31 | 42.77% (159) | siderophore binding protein |
| M. ulcerans Agy99 | MUL_4088 | - | 8e-93 | 91.38% (174) | hypothetical protein MUL_4088 |
| M. vanbaalenii PYR-1 | Mvan_1729 | - | 5e-08 | 37.93% (116) | nucleotidyl transferase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5191|M.smegmatis_MC2_155 ----MPEPLIIPILGHAPQLDPESFVAPN-----------ASVIGQVSLA
TH_1984|M.thermoresistible__bu ----MPEPLIVALGDHAPQLHPESWVAPT-----------ASVIGQVKLA
Mflv_2121|M.gilvum_PYR-GCK ----MAEPLIVSLAGHTPDLHPDSWVAPN-----------ATVIGQVVLA
Mb3555c|M.bovis_AF2122/97 ----MP---LFSFEGRSPRIDPTAFVAPT-----------ATLIGDVTIE
Rv3525c|M.tuberculosis_H37Rv ----MP---LFSFEGRSPRIDPTAFVAPT-----------ATLIGDVTIE
MMAR_5014|M.marinum_M ----MP---LFSFEGRSPRVDPTAFVAPT-----------ATLIGDVTVE
MUL_4088|M.ulcerans_Agy99 ----MP---LFSFEGRSPRVDPTAFVAPT-----------ATLIGDVTVE
MAV_0634|M.avium_104 ----MP---LFAFEGRSPRVDPTAFVAPT-----------ATLIGDVVVE
MAB_4172c|M.abscessus_ATCC_199 ----MP---LYSLDGKSPSVHPEAFVAPT-----------ACLIGDVTVE
MLBr_00753|M.leprae_Br4923 -MATLQVDAVVLVGGKGTRLRPLTLSAPKPMLPTAGLPFLTHLLSRIAAA
Mvan_1729|M.vanbaalenii_PYR-1 MVNPAEVDAVVLVGGMGTRLRPLTLSAPKPMLPTAGLPFLTHLLSRIADA
: . . . : * : **. : ::. :
MSMEG_5191|M.smegmatis_MC2_155 ARASVWYGATLRAESEPIEIGEGS-------NIQDGVTVHVDPGFP----
TH_1984|M.thermoresistible__bu AKVSVWYGATLRAEAEPIDIGEGT-------NIQDGTTIHVDPGFP----
Mflv_2121|M.gilvum_PYR-GCK AGASAWYGAILRAEVEVIDIGAGT-------NIQDGVTIHVDPGFP----
Mb3555c|M.bovis_AF2122/97 AGASVWFNAVLRGDYAPVVVREGA-------NVQDGAVLHAPPGIP----
Rv3525c|M.tuberculosis_H37Rv AGASVWFNAVLRGDYAPVVVREGA-------NVQDGAVLHAPPGIP----
MMAR_5014|M.marinum_M AGASVWFNAVLRGDYAPVVIREGA-------NVQDGSVLHAPPGIP----
MUL_4088|M.ulcerans_Agy99 AGASVWFNAVFRGDYAPVVIREGA-------NVQDGSVLHAPPGIP----
MAV_0634|M.avium_104 AGASVWFNAVLRGDYGPIVVREGA-------NVQDGSVLHAPPGIP----
MAB_4172c|M.abscessus_ATCC_199 AGVSIWFNTVIRADYAPIIIRVGA-------NIQDGSVLHSDPGMP----
MLBr_00753|M.leprae_Br4923 GIEHVILSTSYRDAVFEAEFGDGSKLGLQIDYVIEESPLGTGGGIANVID
Mvan_1729|M.vanbaalenii_PYR-1 GIEHVVLGTSYKANVFESEFGDGSKLGLQIDYVVEDEPLGTGGGIANVAS
. .: : . *: : : : *:.
MSMEG_5191|M.smegmatis_MC2_155 --------------------------------------CRLG--------
TH_1984|M.thermoresistible__bu --------------------------------------VTVG--------
Mflv_2121|M.gilvum_PYR-GCK --------------------------------------VRIG--------
Mb3555c|M.bovis_AF2122/97 --------------------------------------VDIG--------
Rv3525c|M.tuberculosis_H37Rv --------------------------------------VDIG--------
MMAR_5014|M.marinum_M --------------------------------------VDIG--------
MUL_4088|M.ulcerans_Agy99 --------------------------------------VDIG--------
MAV_0634|M.avium_104 --------------------------------------VDIG--------
MAB_4172c|M.abscessus_ATCC_199 --------------------------------------VDIG--------
MLBr_00753|M.leprae_Br4923 QLRHDTVMVFNGDVLSGVDLGQLLGFQRTNFADVTLHLVRVGDPRAFGCV
Mvan_1729|M.vanbaalenii_PYR-1 KLRYDTAVVFNGDVLSGCDLRALLDSHVSRDADVTLHLVRVGDPRAFGCV
:*
MSMEG_5191|M.smegmatis_MC2_155 ------------------------TGVTVGHNVVLHGCT-----------
TH_1984|M.thermoresistible__bu ------------------------AAVSVGHNAVLHGCT-----------
Mflv_2121|M.gilvum_PYR-GCK ------------------------AGVSVGHNAVLHGCT-----------
Mb3555c|M.bovis_AF2122/97 ------------------------PGATVAHLCVIHGVH-----------
Rv3525c|M.tuberculosis_H37Rv ------------------------PGATVAHLCVIHGVH-----------
MMAR_5014|M.marinum_M ------------------------PGATVAHLCVIHGVH-----------
MUL_4088|M.ulcerans_Agy99 ------------------------PGATVAHLCVIHGVH-----------
MAV_0634|M.avium_104 ------------------------PGATVAHLCVIHGAH-----------
MAB_4172c|M.abscessus_ATCC_199 ------------------------EGATVAHMCLVHSCT-----------
MLBr_00753|M.leprae_Br4923 STE-DGRVTAFLEKTQDPPTDQINAGCYVFERNVIDRIPRGREVSVEREV
Mvan_1729|M.vanbaalenii_PYR-1 PTDSDGNVTAFLEKTQDPPTDQINAGCYVFKRSVIDAIPKGRALSVEREI
. * . ::.
MSMEG_5191|M.smegmatis_MC2_155 --------------------------------------------------
TH_1984|M.thermoresistible__bu --------------------------------------------------
Mflv_2121|M.gilvum_PYR-GCK --------------------------------------------------
Mb3555c|M.bovis_AF2122/97 --------------------------------------------------
Rv3525c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_5014|M.marinum_M --------------------------------------------------
MUL_4088|M.ulcerans_Agy99 --------------------------------------------------
MAV_0634|M.avium_104 --------------------------------------------------
MAB_4172c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00753|M.leprae_Br4923 FPTLLSDADVKVCGYVDATYWRDMGTPEDFVRGSADLVRGIAPSPALGGH
Mvan_1729|M.vanbaalenii_PYR-1 FPQLLSDG-KRVCGYVDATYWRDMGTPEDFVRGSADLVRGIAPSPALHGH
MSMEG_5191|M.smegmatis_MC2_155 -----VEDDSLIGMGAVVLNGAVIGAGSLVAA-----GAVVPQGMVVPPR
TH_1984|M.thermoresistible__bu -----VEEGALIGMGAIILNGAVVGAGSLVAA-----GALVPQGFVVPPR
Mflv_2121|M.gilvum_PYR-GCK -----VEENSLVGMGAVVLNGAVVGAGSLIAA-----GAVVPQGAVIPPG
Mb3555c|M.bovis_AF2122/97 -----VGSEALIANHATVLDGAVIGARCMIAA-----GALVVAGTQIPAG
Rv3525c|M.tuberculosis_H37Rv -----VGSEALIANHATVLDGAVIGARCMIAA-----GALVVAGTQIPAG
MMAR_5014|M.marinum_M -----VGAEALIANHATILDGAVIGARCLVAA-----HSLVVGGTQIPEG
MUL_4088|M.ulcerans_Agy99 -----VGAEALIANHATVLDGAVIGARCLVAA-----HSLVVGGTQIPEG
MAV_0634|M.avium_104 -----VGPEALIANHATVLDGAVIGAGSLVAA-----HSLVTAGTQIPAG
MAB_4172c|M.abscessus_ATCC_199 -----VGDGALIGNHATVLDGASIGARSMVAA-----GSLVLGGAQIPAD
MLBr_00753|M.leprae_Br4923 RGEHLVHDGAAVSPGAVLIGGTVVGRGAEIGPGVRLDGAVIFDGAKVEAG
Mvan_1729|M.vanbaalenii_PYR-1 RGESLVHDGASVAPGALLIGGTVVGRGAEVAGGARLDGAVIFDGVKIGAG
* : :. * ::.*: :* . :. ::: * :
MSMEG_5191|M.smegmatis_MC2_155 SLVA----------GVPGKVRREMS-DDEVEHNRFN----AKAYVHLIDL
TH_1984|M.thermoresistible__bu SLVA----------GVPGKVRRELS-DDEVAANRFN----SQVYQGLIEP
Mflv_2121|M.gilvum_PYR-GCK SMVA----------GVPGKVRRQLG-EDELAGIRTN----ATLYQELVKA
Mb3555c|M.bovis_AF2122/97 MLVT----------GAPAKVKGPIEGTGAEMWVNVN----PQAYRDLAAR
Rv3525c|M.tuberculosis_H37Rv MLVT----------GAPAKVKGPIEGTGAEMWVNVN----PQAYRDLAAR
MMAR_5014|M.marinum_M MLVT----------GAPAKVKGPIAGTGAEMWVNVN----PQAYRDLAAR
MUL_4088|M.ulcerans_Agy99 MLVT----------GAPAKVKGPIAGTGAEMWVNVN----PQAYRDLAAR
MAV_0634|M.avium_104 MLAM----------GAPAQIKGPVAGTDAQLWVKLN----PQAYRDLAQR
MAB_4172c|M.abscessus_ATCC_199 VLVM----------GSPAKVKGPVAGTPAEFLTKGV----PAAYRELGKR
MLBr_00753|M.leprae_Br4923 SVIERSILGFGVRIGPRALIRDGVIGDGADIGARCELLRGARVWPGVSIP
Mvan_1729|M.vanbaalenii_PYR-1 AVIERSIIGFGARIGPRALIRDGVIGDGADIGARCELLRGARVWPGITIP
: * . :: : . . : :
MSMEG_5191|M.smegmatis_MC2_155 HRQAG-----
TH_1984|M.thermoresistible__bu HRTANS----
Mflv_2121|M.gilvum_PYR-GCK HRDAYNS---
Mb3555c|M.bovis_AF2122/97 HLAGLEPM--
Rv3525c|M.tuberculosis_H37Rv HLAGLEPM--
MMAR_5014|M.marinum_M HMAGLEPL--
MUL_4088|M.ulcerans_Agy99 HMAGLEPL--
MAV_0634|M.avium_104 HLAGLEPI--
MAB_4172c|M.abscessus_ATCC_199 YLAGLDK---
MLBr_00753|M.leprae_Br4923 DGGIRYSSDV
Mvan_1729|M.vanbaalenii_PYR-1 DGGIRFSTDI