For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LTGPEHGSASTIEILPVIGLPEFRPGDDLSAAVAAAAPWLRDGDVVVVTSKVVSKCEGRLVPAPEDPEQR DRLRRKLIEDEAVRVLARKDRTLITENRLGLVQAAAGVDGSNVGRSELALLPVDPDASAATLRAGLRERL GVTVAVVITDTMGRAWRNGQTDAAVGAAGLAVLRNYAGVRDPYGNELVVTEVAVADEIAAAADLVKGKLT ATPVAVVRGFGVSDDGSTARQLLRPGANDLFWLGTAEALELGRQQAQLLRRSVRRFSTDPVPGDLVEAAV AEALTAPAPHHTRPTRFVWLQTPAIRARLLDRMKDKWRSDLTSDGLPADAIERRVARGQILYDAPEVVIP MLVPDGAHSYPDAARTDAEHTMFTVAVGAAVQALLVALAVRGLGSCWIGSTIFAADLVRDELDLPVDWEP LGAIAIGYADEPSGLRDPVPAADLLILK
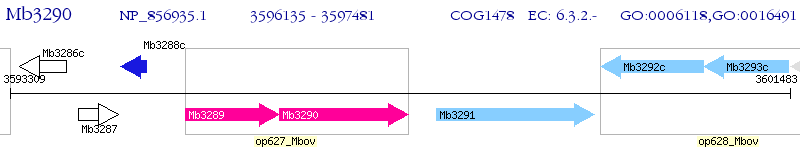
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3290 | fbiB | - | 100% (448) | F420-0--gamma-glutamyl ligase |
| M. bovis AF2122 / 97 | Mb0314 | - | 3e-06 | 27.68% (177) | putative oxidoreductase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4731 | - | 0.0 | 74.21% (442) | F420-0--gamma-glutamyl ligase |
| M. tuberculosis H37Rv | Rv3262 | fbiB | 0.0 | 100.00% (448) | F420-0--gamma-glutamyl ligase |
| M. leprae Br4923 | MLBr_00758 | - | 0.0 | 82.25% (445) | F420-0--gamma-glutamyl ligase |
| M. abscessus ATCC 19977 | MAB_3608 | - | 1e-173 | 67.11% (447) | F420-0--gamma-glutamyl ligase |
| M. marinum M | MMAR_1280 | fbiB | 0.0 | 82.66% (444) | F420 biosynthesis protein FbiB |
| M. avium 104 | MAV_4225 | - | 0.0 | 84.68% (444) | F420-0--gamma-glutamyl ligase |
| M. smegmatis MC2 155 | MSMEG_1829 | - | 0.0 | 79.64% (447) | F420-0--gamma-glutamyl ligase |
| M. thermoresistible (build 8) | TH_0777 | - | 0.0 | 76.35% (444) | PROBABLE F420 BIOSYNTHESIS PROTEIN FBIB |
| M. ulcerans Agy99 | MUL_2607 | fbiB | 0.0 | 82.21% (444) | F420-0--gamma-glutamyl ligase |
| M. vanbaalenii PYR-1 | Mvan_1732 | - | 0.0 | 74.43% (442) | F420-0--gamma-glutamyl ligase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3290|M.bovis_AF2122/97 MTG---------PEHGSASTIEILPVIGLPEFRPGDDLSAAVAAAAPWLR
Rv3262|M.tuberculosis_H37Rv MTG---------PEHGSASTIEILPVIGLPEFRPGDDLSAAVAAAAPWLR
MLBr_00758|M.leprae_Br4923 MTSSDSHRSAPSPEHGTASTIEILPVAGLPEFRPGDDLSAALAAAAPWLR
MMAR_1280|M.marinum_M MSGPAL---PPTGEHGTASAIEILPVEGLPEFRPGDELSAAIATAAPWLR
MUL_2607|M.ulcerans_Agy99 MSGPAL---PPTGEHGTVSAIEILPVEGLPEFRPGNELSAAIATAAPWLR
MAV_4225|M.avium_104 MS--------PAGEHGTAAPIEILPVAGLPEFRPGDDLGAAVAKAAPWLR
Mflv_4731|M.gilvum_PYR-GCK -----------MTDHGTSARVEVLPVPGLPEFRPGDDLAAAIADAAPWLR
Mvan_1732|M.vanbaalenii_PYR-1 -----------MTDHGTSARVELLPVPGLPEFRPGDNLSAAIAEAAPWLQ
TH_0777|M.thermoresistible__bu VSTAGGAGQTGSGDHGTAAAIELFPVTGLPQFCPGDDLTAAVAAAAPWLR
MSMEG_1829|M.smegmatis_MC2_155 ------MSAAANAEHGAADRVEILPVPGLPEFRPGDDLVGSLAEAAPWLR
MAB_3608|M.abscessus_ATCC_1997 MTPPADS---PAPEHGSAAPVQILPVTGLPEFRPGDDVAAAIAGAAPWLR
:**: ::::** ***:* **::: .::* *****:
Mb3290|M.bovis_AF2122/97 DGDVVVVTSKVVSKCEGRLVPAPEDPEQRDRLRRKLIEDEAVRVLARKDR
Rv3262|M.tuberculosis_H37Rv DGDVVVVTSKVVSKCEGRLVPAPEDPEQRDRLRRKLIEDEAVRVLARKDR
MLBr_00758|M.leprae_Br4923 DGDVVVVTSKVVSKCEGRLVPAPEDTRGRNELRRKLINDETIRVLARKGR
MMAR_1280|M.marinum_M DGDVVVVTSKVVSKCEGRLVTAPADEEARDQLRRKLVDDEAVRVLARKGR
MUL_2607|M.ulcerans_Agy99 DGDVVVVTSKVVSKCEGRLVTAPADEEARDQLRRKLVDDEAVRVLARKGR
MAV_4225|M.avium_104 DGDVVVVTSKAVSKCEGRLVPAPADPEERDRLRRKLVDEEAVRVLARKGR
Mflv_4731|M.gilvum_PYR-GCK DDDVVVVTSKVLSKTEGRMVEAPTDPDERDALRRKLIDGEAVRVLARKGR
Mvan_1732|M.vanbaalenii_PYR-1 DDDVLVVTSKVLSKTEGRMVAAPADPEERDALRRKLIDSEAVRVLARKGR
TH_0777|M.thermoresistible__bu DNDVVVVTSKVVSKCEGRIIAAPADPEQRDALRRKLIDEEAVRVLARKGR
MSMEG_1829|M.smegmatis_MC2_155 DGDVLVVTSKVVSKCEGRIVAAPSDPEERDTLRRKLIDDEAVRVLARKGR
MAB_3608|M.abscessus_ATCC_1997 DGDVIVITSKIISKAEGRIVAAPTDPEARDTLRRKLIDSESIRVLARKGK
*.**:*:*** :** ***:: ** * *: *****:: *::******.:
Mb3290|M.bovis_AF2122/97 TLITENRLGLVQAAAGVDGSNVGRSELALLPVDPDASAATLRAGLRERLG
Rv3262|M.tuberculosis_H37Rv TLITENRLGLVQAAAGVDGSNVGRSELALLPVDPDASAATLRAGLRERLG
MLBr_00758|M.leprae_Br4923 TLIIENGLGLVQAAAGVDGSNVGRGELALLPVNPDASAAVLRIGLRAMLG
MMAR_1280|M.marinum_M TLITESRLGLVQAAAGVDGSNVDRSELALLPVDPDASAAALRTGLRERLG
MUL_2607|M.ulcerans_Agy99 TLITESRLGLVQAAAGVDGSNVDRSELALLPVDPDASAAALRTGLRERLG
MAV_4225|M.avium_104 TLITENRHGLVQAAAGVDGSNVGRDELALLPLDPDASAAALRARLRELLG
Mflv_4731|M.gilvum_PYR-GCK TLITENALGLVQAAAGVDGSNVDSNELALLPVDPDGSAAELVAALHERLG
Mvan_1732|M.vanbaalenii_PYR-1 TLITENALGLVQAAAGVDGSNVESNELALLPVDPDGSAAALVAALRERLG
TH_0777|M.thermoresistible__bu TLITENALGLVQAAAGVDGSNVAATELALLPVDPDASAAALRAGLRQRLG
MSMEG_1829|M.smegmatis_MC2_155 TLITENAIGLVQAAAGVDGSNVGSTELALLPVDPDRSAATLREGLRERLG
MAB_3608|M.abscessus_ATCC_1997 TLITENTIGIVQAAAGIDASNVDTAELVLLPTDPDASAAAVRLALSQRLG
*** *. *:******:*.*** **.*** :** *** : * **
Mb3290|M.bovis_AF2122/97 VTVAVVITDTMGRAWRNGQTDAAVGAAGLAVLRNYAGVRDPYGNELVVTE
Rv3262|M.tuberculosis_H37Rv VTVAVVITDTMGRAWRNGQTDAAVGAAGLAVLRNYAGVRDPYGNELVVTE
MLBr_00758|M.leprae_Br4923 VNVAVVITDTMGRAWRNGQTDVAIGAAGLAVLHNYSGAVDRYGNELVVTE
MMAR_1280|M.marinum_M VTVAVVITDTMGRAWRNGQIDAAVGAAGVSVLHNYSGATDRHGNELIVTE
MUL_2607|M.ulcerans_Agy99 VTVAVVITDTMGRAWRNGQIDAAVGAAGVSVLHNYSGATDRHGNELIVTE
MAV_4225|M.avium_104 VEVAVLVTDTMGRAWRNGQTDAAVGAAGLAVLHGYSGAVDQHGNELLVTE
Mflv_4731|M.gilvum_PYR-GCK VRVAVVVTDTMGRAWRTGQTDVAIGAAGLTVLHAYAGEHDQHGNELIVTE
Mvan_1732|M.vanbaalenii_PYR-1 VRIGVVVTDTMGRAWRTGQTDVAIGAAGLTVLHAYAGERDQHGNELIVTE
TH_0777|M.thermoresistible__bu VDVGVIITDTMGRAWRTGQTDFAIGAAGLTVLHGYAGSHDRHGNELFVTE
MSMEG_1829|M.smegmatis_MC2_155 VTVGVVITDTMGRAWRTGQTDFAIGASGLTVLQGYAGSRDRHGNELVVTE
MAB_3608|M.abscessus_ATCC_1997 VKVAVVITDTMGRAWRNGQTDAAIGSAGIPVLYGYAGAKDKHGNELQVTE
* :.*::*********.** * *:*::*:.** *:* * :**** ***
Mb3290|M.bovis_AF2122/97 VAVADEIAAAADLVKGKLTATPVAVVRGFGVSDDGSTARQLLRPGANDLF
Rv3262|M.tuberculosis_H37Rv VAVADEIAAAADLVKGKLTATPVAVVRGFGVSDDGSTARQLLRPGANDLF
MLBr_00758|M.leprae_Br4923 IAVADEVAAATDLVKGKLTAMPVAVVRGLSPTDDGSTAQHLLRNGPDDLF
MMAR_1280|M.marinum_M IAVADEIAAAADLVKGKLTAMPVAVVRGLPVRDDGSTARNLVRPGEEDLF
MUL_2607|M.ulcerans_Agy99 IAVADEIAAAADLVKGKLTAMPVAVVRGLPVRDDGSTARNLVRPGEEDLF
MAV_4225|M.avium_104 VAIADEIAAAADLVKGKLTAMPVAVVRGLSVTDDGSTARQLLRPGTEDLF
Mflv_4731|M.gilvum_PYR-GCK IAVADEIAAAADLVKGKLTAVPVAVVRGLAVRDDGSSARNLVRAGEEDLF
Mvan_1732|M.vanbaalenii_PYR-1 IAVADEIAAAADLVKGKLTAVPVAVVRGLSLRDDGSSARNLVRAGEEDLF
TH_0777|M.thermoresistible__bu VAVADEIAAAADLVKGKLTDVPVAVVRGLTLRDDGSTARTLVRPGQDDLF
MSMEG_1829|M.smegmatis_MC2_155 VAVADEIAAAADLVKGKLTAIPVAVVRGLRLPDDGSTAHRLVRAGEDDLF
MAB_3608|M.abscessus_ATCC_1997 VAIADEISAAADLVKGKLTDVPVAVVRGLTLLDDGSAASDLLRSGPDDLF
:*:***::**:******** *******: ****:* *:* * :***
Mb3290|M.bovis_AF2122/97 WLGTAEALELGRQQAQLLRRSVRRFSTDPVPGDLVEAAVAEALTAPAPHH
Rv3262|M.tuberculosis_H37Rv WLGTAEALELGRQQAQLLRRSVRRFSTDPVPGDLVEAAVAEALTAPAPHH
MLBr_00758|M.leprae_Br4923 WLGTTEALELGRQQAQLLRRSVRQFSDEPIAAELIETAVAEALTAPAPHH
MMAR_1280|M.marinum_M WLGTAEAIELGHQQAQLMRRSIRQFANEPVPPQLLEAAVAEALTAPAPHH
MUL_2607|M.ulcerans_Agy99 WLGTAEAIELGHQQAQLMRRSIRQFADEPVPPQLLEAAVAEALTAPAPHH
MAV_4225|M.avium_104 WLGTAEAIELGRRQAQLLRRSVRRFSAEPVPAELVREAVAEALTAPAPHH
Mflv_4731|M.gilvum_PYR-GCK WLGTDEALALGRSQAQLLRRSVRQFSPEPVDGALIGAAVGEALTAPAPHH
Mvan_1732|M.vanbaalenii_PYR-1 WLGTEEALALGRSQAQLLRRSVRTFNSEPVPGDVIEAAVGEALTAPAPHH
TH_0777|M.thermoresistible__bu WLGTEEAIRLGRSQAQLLRRSVRRFDSEPVPADLIEAAVAEALTAPAPHH
MSMEG_1829|M.smegmatis_MC2_155 WLGTAEAIELGRRQAQLLRRSVRRFSAEPVPHDAIEAAVGEALTAPAPHH
MAB_3608|M.abscessus_ATCC_1997 WLGTNEALEQGRREAILVRRSIRQFADIPVDPDLLREAIGEALTAPAPHH
**** **: *: :* *:***:* * *: : *:.**********
Mb3290|M.bovis_AF2122/97 TRPTRFVWLQTPAIRARLLDRMKDKWRSDLTSDGLPADAIERRVARGQIL
Rv3262|M.tuberculosis_H37Rv TRPTRFVWLQTPAIRARLLDRMKDKWRSDLTSDGLPADAIERRVARGQIL
MLBr_00758|M.leprae_Br4923 TRPVRFVWLQTPAVRTRLLDRMADKWRLDLASDALPADAIAQRVARGQIL
MMAR_1280|M.marinum_M TRPVRFVWLQSHATRIALLDAMKDKWRHDLAGDGRSADSIERRVTRGQIL
MUL_2607|M.ulcerans_Agy99 TRPVRFVWLQSHATRIALLDAMKDKWRHDLVGDGRSADSIERRVTRGQIL
MAV_4225|M.avium_104 TRPVRFVWLQTPAVRTRLLDAMKDKWRADLAGDGRPAESIERRVARGQIL
Mflv_4731|M.gilvum_PYR-GCK TRPVRFVWVQDGALRVGLLDEMKERWRADLLADGRDAESVERRVSRGQIL
Mvan_1732|M.vanbaalenii_PYR-1 TRPVRFVWVQDASVRVGLLDKMKDRWRADLLGDGRDPESVERRVNRGQIL
TH_0777|M.thermoresistible__bu TRPVRFVWLQNWTARTRLLDRMKDRWRADLAGDGASPEAVERRVGRGQIL
MSMEG_1829|M.smegmatis_MC2_155 TRPVRFVWVQDSETRTRLLDRMKEQWRADLTADGLDADAVDRRVARGQIL
MAB_3608|M.abscessus_ATCC_1997 THPVRFVWVRDLTTRRALLDALKQAWAADLTADGRSPESVQKRVARGQIL
*:*.****:: * *** : : * ** .*. .::: :** *****
Mb3290|M.bovis_AF2122/97 YDAPEVVIPMLVPDGAHSYPDAARTDAEHTMFTVAVGAAVQALLVALAVR
Rv3262|M.tuberculosis_H37Rv YDAPEVVIPMLVPDGAHSYPDAARTDAEHTMFTVAVGAAVQALLVALAVR
MLBr_00758|M.leprae_Br4923 YDAPEVIIPFMVPDGAHAYPDAARASAEHTMFIVAVGAAVQALLVALAVR
MMAR_1280|M.marinum_M YDAPEVVIPMLVPDGAHTYPDGARTQAEHTMFTVAVGAAVQALLVALAVR
MUL_2607|M.ulcerans_Agy99 YDAPEVVIPMLVPDGAHTYPDGARTQAEHTMFTVAVGAAVQALLVALAVR
MAV_4225|M.avium_104 YDAPEVVIPMLVPDGAHSYPDAARTDAEHTMFTVAVGAAVQALLVGLAVR
Mflv_4731|M.gilvum_PYR-GCK YDAPELIIPFMVPDGAHSYPDERRTAAEHTMFTVAAGAAVQGLLVALAVR
Mvan_1732|M.vanbaalenii_PYR-1 YDAPELVIPFMVPDGAHSYPDARRTAAEHTMFTVAAGAAVQGLLVALAAR
TH_0777|M.thermoresistible__bu YDAPELIVPFLVADGAHHYPDARRAAAEHTMFTVAGGAAVQALLVALAVR
MSMEG_1829|M.smegmatis_MC2_155 YDAPELVIPFLVPDGAHSYPDDARTAAEHTMFTVAVGAAVQGLLVALAVR
MAB_3608|M.abscessus_ATCC_1997 YDAPEIVIPFLVPDGAHDYPDDRRTAAERTMFTVAVGAAVQALLVALAAR
*****:::*::*.**** *** *: **:*** ** *****.***.**.*
Mb3290|M.bovis_AF2122/97 GLGSCWIGSTIFAADLVRDELDLPVDWEPLGAIAIGYADE----PSGLRD
Rv3262|M.tuberculosis_H37Rv GLGSCWIGSTIFAADLVRDELDLPVDWEPLGAIAIGYADE----PSGLRD
MLBr_00758|M.leprae_Br4923 GLGSCWIGSTIFADDLVRAELELPADWEPLGAIAIGYAHE----PTDLRE
MMAR_1280|M.marinum_M GLGSCWIGSTIFAADLVRAQLQLPADWEPLGAIAIGYAQQ----PAELRD
MUL_2607|M.ulcerans_Agy99 GLGSCWIGSTIFAADLVRAQLQLPADWEPLGAIAIGYAQQ----PAELRD
MAV_4225|M.avium_104 GLGSCWIGSTIFAADLVRAVLGLPADWEPLGAIAIGYAAE----PAGPRD
Mflv_4731|M.gilvum_PYR-GCK GVGSCWIGSTIFAPELVRAALEVPGDWEPLGAIAIGYGTD----PVTPRE
Mvan_1732|M.vanbaalenii_PYR-1 SVGSCWIGSTIFAPDLVRAELGVPDDWEPLGAIAIGYPEA----PLAPRH
TH_0777|M.thermoresistible__bu EIGSCWVGSTIFAPEVVRTELGLPDDWEPLGAVAIGYPAE----PAGPRD
MSMEG_1829|M.smegmatis_MC2_155 DIGSCWIGSTIFAADLVRAELELPDDWEPLGAIAIGYPEQTPQ-PLGPRD
MAB_3608|M.abscessus_ATCC_1997 QVGSCWIGSTIFAGDVVRKQLVVDSSWEPMGAIAIGYPHGYPEEGLDPRT
:****:****** ::** * : .***:**:**** *
Mb3290|M.bovis_AF2122/97 PVPAADLLILK
Rv3262|M.tuberculosis_H37Rv PVPAADLLILK
MLBr_00758|M.leprae_Br4923 PVRVADLLLRK
MMAR_1280|M.marinum_M PVHPGDLLIRK
MUL_2607|M.ulcerans_Agy99 PVHPGDLLIRK
MAV_4225|M.avium_104 PADPGDLLIRK
Mflv_4731|M.gilvum_PYR-GCK PVPTDGLLVRR
Mvan_1732|M.vanbaalenii_PYR-1 PVPTDDLLVRR
TH_0777|M.thermoresistible__bu PAPTEGLLVFR
MSMEG_1829|M.smegmatis_MC2_155 PVPTDELLVRK
MAB_3608|M.abscessus_ATCC_1997 PLPPGQMLVEL
* :*: