For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VMRFLDGHPPGYDLTYNDVFIVPNRSEVASRFDVDLSTADGSGTTIPVVVANMTAVAGRRMAETVARRGG IVILPQDLPIPAVKQTVAFVKSRDLVLDTPVTLAPDDSVSDAMALIHKRAHGVAVVILEGRPIGLVRESS CLGVDRFTRVRDIAVTDYVTAPAGTEPRKIFDLLEHAPVDVAVLTDADGTLAGVLSRTGAIRAGIYTPAT DSAGRLRIGAAVGINGDVGAKARALAEAGVDVLVIDTAHGHQVKTLDAIKAVSALDLGLPLAAGNVVSAE GTRDLLKAGANVVKVGVGPGAMCTTRMMTGVGRPQFSAVLECASAARQLGGHIWADGGIRHPRDVALALA AGASNVMIGSWFAGTYESPGDLMRDRDDQPYKESYGMASKRAVVARTGADNPFDRARKALFEEGISTSRM GLDPDRGGVEDLIDHITSGVRSTCTYVGASNLAELHERAVVGVQSGAGFAEGHPLPAGW
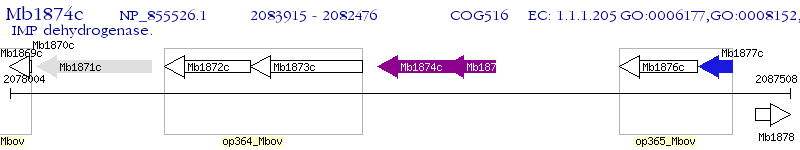
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1874c | guaB1 | - | 100% (479) | inosine 5-monophosphate dehydrogenase |
| M. bovis AF2122 / 97 | Mb3445c | guaB2 | 2e-65 | 35.98% (478) | inosine 5'-monophosphate dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3360 | - | 0.0 | 82.01% (478) | inositol-5-monophosphate dehydrogenase |
| M. tuberculosis H37Rv | Rv1843c | guaB1 | 0.0 | 100.00% (479) | inosine 5-monophosphate dehydrogenase |
| M. leprae Br4923 | MLBr_02066 | guaB1 | 0.0 | 86.40% (478) | inosine 5-monophosphate dehydrogenase |
| M. abscessus ATCC 19977 | MAB_2412c | - | 0.0 | 75.10% (478) | inosine 5-monophosphate dehydrogenase |
| M. marinum M | MMAR_2717 | guaB1 | 0.0 | 89.12% (478) | inosine-5'-monophosphate dehydrogenase GuaB1 |
| M. avium 104 | MAV_2872 | - | 0.0 | 86.61% (478) | inositol-5-monophosphate dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_3634 | - | 0.0 | 84.31% (478) | inositol-5-monophosphate dehydrogenase |
| M. thermoresistible (build 8) | TH_2384 | - | 0.0 | 80.21% (480) | IMP dehydrogenase family protein |
| M. ulcerans Agy99 | MUL_3036 | guaB1 | 0.0 | 88.91% (478) | inosine 5-monophosphate dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_3090 | - | 0.0 | 84.10% (478) | inositol-5-monophosphate dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3360|M.gilvum_PYR-GCK -MQFLDGHRPPYDLTYNDVFVVPGRSEVTSRFDVDLSTVDGAGTTIPVVV
Mvan_3090|M.vanbaalenii_PYR-1 -MKFLDDQRPPYDLTYNDVFVVPGRSEVLSRFDVDLSTVDGSGTTIPVVV
Mb1874c|M.bovis_AF2122/97 MMRFLDGHPPGYDLTYNDVFIVPNRSEVASRFDVDLSTADGSGTTIPVVV
Rv1843c|M.tuberculosis_H37Rv MMRFLDGHPPGYDLTYNDVFIVPNRSEVASRFDVDLSTADGSGTTIPVVV
MLBr_02066|M.leprae_Br4923 -MRFLDGHQPRFDLTYKDVFIVPHRSDVASRFDVDLSTYDGSGTTIPVVV
MAV_2872|M.avium_104 -MRFLDGHRPGYDLTYNDVFIMPNRSEVPSRFDVDLSTGDGSGTTIPVVV
MMAR_2717|M.marinum_M -MRFLDGHRPGYDLTYNDVFIMPNRSLVASRFDVDLSTADGSGTTIPVVV
MUL_3036|M.ulcerans_Agy99 -MRFLDGHRPGYDLTYNDVFIMPNRSLVASRFDVDLSTADGSGTTIPVVV
MSMEG_3634|M.smegmatis_MC2_155 -MRFLDGHTPAYDLTYNDVFVVPGRSDVASRFDVDLSTVDGSGTTIPVVV
TH_2384|M.thermoresistible__bu -MRFLDGHRPPYDLTYDDVFIMPGRSDLASRLDVDLSTVDGSGTTIPIVV
MAB_2412c|M.abscessus_ATCC_199 -MKFLDGQRPPYDLTYDDVFVVPNHSDVTSRFDVDLTTADGTGTTIPIVV
*:***.: * :****.***::* :* : **:****:* **:*****:**
Mflv_3360|M.gilvum_PYR-GCK ANMTAVAGRRMAETVARRGGIVVLPQDLPPSAVRHTVDFVKSRDTVVDTP
Mvan_3090|M.vanbaalenii_PYR-1 ANMTAVAGRRMAETVARRGGIVVLPQDLPVSAVKQTVDFIKSRDTVVDTP
Mb1874c|M.bovis_AF2122/97 ANMTAVAGRRMAETVARRGGIVILPQDLPIPAVKQTVAFVKSRDLVLDTP
Rv1843c|M.tuberculosis_H37Rv ANMTAVAGRRMAETVARRGGIVILPQDLPIPAVKQTVAFVKSRDLVLDTP
MLBr_02066|M.leprae_Br4923 ANMTTVAGQRMAETVARRGGLVILPQDLPIATVQQAVEFVKSRDLVIDTP
MAV_2872|M.avium_104 ANMTAVAGRRMAETVARRGGLVILPQDLPITAVQQTVQFVKSRDLVLDTP
MMAR_2717|M.marinum_M ANMTAVAGRRMAETVARRGGIVVLPQDLPIPAVHQTVEFIKSRDLVLDTP
MUL_3036|M.ulcerans_Agy99 ANMTAVAGRRMAETVARRGGIVVLPQDLPIPAVHQTVEFIKSRDLVLDTP
MSMEG_3634|M.smegmatis_MC2_155 ANMTAVAGRRMAETVARRGGIVVLPQDLPITAVSETVDFVKSRDLVVDTP
TH_2384|M.thermoresistible__bu ANMTAVAGRRMAETVARRGGLVVLPQDLPIEVVRHTVDFVKSRDLVVDTP
MAB_2412c|M.abscessus_ATCC_199 ANMTAVAGRRMAETVARRGGITVLPQDLPVQAVAQTVEFIKSRDLVYDTP
****:***:***********:.:****** .* .:* *:**** * ***
Mflv_3360|M.gilvum_PYR-GCK VTLSPDDSVSDAAALIHKRAHGAAVVLDHD-RPVGLVTEAACHGVDRFTR
Mvan_3090|M.vanbaalenii_PYR-1 VTLSPDDSVSDAIALIHKRAHGAAVVLFEN-RPIGLVTEASCAGVDRFTR
Mb1874c|M.bovis_AF2122/97 VTLAPDDSVSDAMALIHKRAHGVAVVILEG-RPIGLVRESSCLGVDRFTR
Rv1843c|M.tuberculosis_H37Rv VTLAPDDSVSDAMALIHKRAHGVAVVILEG-RPIGLVRESSCLGVDRFTR
MLBr_02066|M.leprae_Br4923 VLLTPNDSVSDATTLIHKRAHGVAVVAFEG-RPIGLVRESSCLGVDRFTR
MAV_2872|M.avium_104 VLLEPDDSVSDAMALIHKRAHGVAVVAFEG-RPLGLVSESSCLGVDRFTR
MMAR_2717|M.marinum_M VTLAPEDSVSDAIALIHKRAHGVAVVVSEG-RPIGLVTEASCVGVDRFAR
MUL_3036|M.ulcerans_Agy99 VTLAPEDSVSDAIALIHKRAHGVAVVVSEG-RPIGLVTEASCVGVDRFAR
MSMEG_3634|M.smegmatis_MC2_155 VTLSPEDSVSDANALLHKRAHGAAVVVFEG-RPIGLVTEANCAGVDRFAR
TH_2384|M.thermoresistible__bu VTLGPEDSVSDAIALIHKRAHGAAVVVDEHNRPVGLVTEDNCVGVDRFTR
MAB_2412c|M.abscessus_ATCC_199 VTLAPDDSAGEAIALIHKRSHGAAVVLEHG-KPLGLVTEAACTGIDRFTR
* * *:**..:* :*:***:**.*** . :*:*** * * *:***:*
Mflv_3360|M.gilvum_PYR-GCK VCDVAITDFVTAPAGTDPRKVFDLLEHA-HIDVAVLTDADGSLAGVLTRT
Mvan_3090|M.vanbaalenii_PYR-1 VRDVVVSDFVSVPVGTDPRKVFDLLEHA-PVDVAVLTEADGSLAGVLTRT
Mb1874c|M.bovis_AF2122/97 VRDIAVTDYVTAPAGTEPRKIFDLLEHA-PVDVAVLTDADGTLAGVLSRT
Rv1843c|M.tuberculosis_H37Rv VRDIAVTDYVTAPAGTEPRKIFDLLEHA-PVDVAVLTDADGTLAGVLSRT
MLBr_02066|M.leprae_Br4923 VRDVATTDFVTAPVGTDPRKIFDLLEHA-PIDVAVLTNADGTLAGVLTRT
MAV_2872|M.avium_104 VRDVAMTDFVVAPVGTEPRKIFDLLEQA-PIGVAVVTNADGTLAGVLTRT
MMAR_2717|M.marinum_M VRDIAALDFVSAPVGTEPRKVFDLLEHS-PIEVAVLTRPDGTLAGVLSRT
MUL_3036|M.ulcerans_Agy99 VRDIAALDFVSAPVGTEPRKVFDLLEHS-PIEVAVLTRPDGTLAGVLSRT
MSMEG_3634|M.smegmatis_MC2_155 VRDIALSDFVTAPVGTDPREVFDLLEHA-PIDVAVMTAPDGTLAGVLTRT
TH_2384|M.thermoresistible__bu VREVAVGEFVSAPVGTAPRKVFDLIEHAQPFDLAVMTASDGTLAGVLTRR
MAB_2412c|M.abscessus_ATCC_199 VHDIAATDFVTVPTGTDPRAIFDRLSEA-HVPVAVLTRPDGTMAGILTRT
* ::. ::* .*.** ** :** :..: . :**:* .**::**:*:*
Mflv_3360|M.gilvum_PYR-GCK GAVRAGIYTPAVDAHGRLRIATAVGINGDVAAKARDLVEAGADLLVIDTA
Mvan_3090|M.vanbaalenii_PYR-1 GAVRAGIYTPAVDADGRLRIAAAVGINGDVGAKARDLAEAGVDVLVIDTA
Mb1874c|M.bovis_AF2122/97 GAIRAGIYTPATDSAGRLRIGAAVGINGDVGAKARALAEAGVDVLVIDTA
Rv1843c|M.tuberculosis_H37Rv GAIRAGIYTPATDSAGRLRIGAAVGINGDVGAKARALAEAGVDVLVIDTA
MLBr_02066|M.leprae_Br4923 GAIRAGIYTPATDARGRLRIGAAVGINGDVAGKAQSLAEAGVDVLVVDTA
MAV_2872|M.avium_104 GAVRAGLYTPAVDAQGRLRVGAAIGINGDVGAKARSLVEAGIDLLVIDTA
MMAR_2717|M.marinum_M GAIRAGIYTPATDAAGRLRIAAAVGINGDVAAKAGALAESGVDLLVIDTA
MUL_3036|M.ulcerans_Agy99 GAIRAGIYTPATDAAGRLRIAAAVGINGDVAAKAGALAESGVDLLVIDTA
MSMEG_3634|M.smegmatis_MC2_155 GAIRAGIYTPAVDAKGRLRIAAAVGINGDVGAKAQALAEAGADLLVIDTA
TH_2384|M.thermoresistible__bu GALRAEIYTPATDAQGRLRVAAAVGINGDVAAKAKALADTGIDLLVIDTA
MAB_2412c|M.abscessus_ATCC_199 GAIRAGLYTPAVDDNGRLRVAGAVGINGDVRAKASALVQAGVDLLVIDTA
**:** :****.* ****:. *:****** .** *.::* *:**:***
Mflv_3360|M.gilvum_PYR-GCK HGHQRKMLDAIAAVAAADLGVPLAAGNVVSATGTRDLINAGATIVKVGVG
Mvan_3090|M.vanbaalenii_PYR-1 HGHQQKMLDTVEAVAALELGLPLVAGNVVSAEGTRDLISAGASIVKVGVG
Mb1874c|M.bovis_AF2122/97 HGHQVKTLDAIKAVSALDLGLPLAAGNVVSAEGTRDLLKAGANVVKVGVG
Rv1843c|M.tuberculosis_H37Rv HGHQVKTLDAIKAVSALDLGLPLAAGNVVSAEGTRDLLKAGANVVKVGVG
MLBr_02066|M.leprae_Br4923 HGYQVKTLEAIKCVASLNLGVPLVAGNVVSAEGTRELLNAGATIVKVGVG
MAV_2872|M.avium_104 HGHQVKTLEAIRTVASLELGVPLVAGNVVSADGTRDLLGAGANIVKVGVG
MMAR_2717|M.marinum_M HGHQVKTLDAISAVASLDLGLPLAAGNVVSAEGTRDLLGAGADIVKVGVG
MUL_3036|M.ulcerans_Agy99 HGHQVKTLDAISAVASLDLGLPLAAGNVVSAEGTRDLLGAGADIVKVGVG
MSMEG_3634|M.smegmatis_MC2_155 HGHQAKMLDAIKAVASLDLGLPLVAGNVVSAEGTRDLIEAGASIVKVGVG
TH_2384|M.thermoresistible__bu HGHQIRMVEAVRAVAALGLGLPLVAGNVVSAEGTRDLIEAGASIVKVGVG
MAB_2412c|M.abscessus_ATCC_199 HGHQQKMLDVLAAVAELSLGVPIAAGNVVSAQGTRDLIRAGADIVKVGVG
**:* : ::.: *: **:*:.******* ***:*: *** :******
Mflv_3360|M.gilvum_PYR-GCK PGAMCTTRMMTGVGRPQFSAVVECASAAKELGGHVWADGGVRHPRDVALA
Mvan_3090|M.vanbaalenii_PYR-1 PGAMCTTRMMTGVGRPQFSAVVECAAAARELGGHVWADGGVRHPRDVALA
Mb1874c|M.bovis_AF2122/97 PGAMCTTRMMTGVGRPQFSAVLECASAARQLGGHIWADGGIRHPRDVALA
Rv1843c|M.tuberculosis_H37Rv PGAMCTTRMMTGVGRPQFSAVLECASAARQLGGHIWADGGIRHPRDVALA
MLBr_02066|M.leprae_Br4923 PGAMCTTRMMTGVGRPQFSAVLECASAARKLNRHVWADGGVRHPRDVALA
MAV_2872|M.avium_104 PGAMCTTRMMTGVGRPQFSAVVECASAARELGGHVWADGGIRHPRDVALA
MMAR_2717|M.marinum_M PGAMCTTRMMTGVGRPQFSAVLECSAAARELGGHVWADGGIRHPRDVALA
MUL_3036|M.ulcerans_Agy99 PGAMCTTRMMTGVGRPQFSAVLECSAAARELGGHVWADGGIRHPRDVALA
MSMEG_3634|M.smegmatis_MC2_155 PGAMCTTRMMTGVGRPQFSAVVECAAAARQLGGHVWADGGVRHPRDVALA
TH_2384|M.thermoresistible__bu PGAMCTTRMMTGVGRPQFSAVLECAAAARELGAHVWADGGVRHPRDVALA
MAB_2412c|M.abscessus_ATCC_199 PGAMCTTRMMTGVGRPQLSAVIECAAAARELGAHVWADGGVRHPRDVALA
*****************:***:**::**::*. *:*****:*********
Mflv_3360|M.gilvum_PYR-GCK LAAGASNVMIGSWFAGTYESPGDLMHDRDGLPYKESYGMASKRAVAARTA
Mvan_3090|M.vanbaalenii_PYR-1 IAAGASNVMIGSWFAGTYESPGDLMRDRDDQPYKESYGMASKRAVAARTA
Mb1874c|M.bovis_AF2122/97 LAAGASNVMIGSWFAGTYESPGDLMRDRDDQPYKESYGMASKRAVVARTG
Rv1843c|M.tuberculosis_H37Rv LAAGASNVMIGSWFAGTYESPGDLMRDRDDQPYKESYGMASKRAVVARTG
MLBr_02066|M.leprae_Br4923 LAAGASNVMIGSWFAGTYESPGDLMRDRHDQPYKESYGMASKRAVVARTV
MAV_2872|M.avium_104 LAAGASNVMIGSWFAGTYESPGDLMRDREDQPYKESYGMASKRAVVARSG
MMAR_2717|M.marinum_M LAAGASNVMIGSWFAGTYESPGDLMRDREDQPYKESYGMASKRAVVARTT
MUL_3036|M.ulcerans_Agy99 LAAGASNVMIGSWFAGTYESPGDLMRDREDQPYKESYGMASKRAVVARTT
MSMEG_3634|M.smegmatis_MC2_155 LAAGASNVMIGSWFAGTYESPGDLLFDRDDRPYKESYGMASKRAVAARTA
TH_2384|M.thermoresistible__bu LAAGASNVMIGSWFAGTYESPGDLMRDRDDRPYKESYGMASKRAVTARTA
MAB_2412c|M.abscessus_ATCC_199 LAAGAANVMIGSWFAGTHESPGDLQLSPTGRRYKDSFGMASKRAVAARTA
:****:***********:****** . . **:*:********.**:
Mflv_3360|M.gilvum_PYR-GCK GDSAFDRARKGLFEEGISTSRMDLDPVRGGVEDLLDHITSGVRSTCTYVG
Mvan_3090|M.vanbaalenii_PYR-1 GDSSFDRARKALFEEGISSSRMYLDPVRGGVEDLIDHITSGVRSTCTYVG
Mb1874c|M.bovis_AF2122/97 ADNPFDRARKALFEEGISTSRMGLDPDRGGVEDLIDHITSGVRSTCTYVG
Rv1843c|M.tuberculosis_H37Rv ADNPFDRARKALFEEGISTSRMGLDPDRGGVEDLIDHITSGVRSTCTYVG
MLBr_02066|M.leprae_Br4923 ADSSFDRARKALFDEGISTSRMGLDPDHGGVEDLIDHITSGVRSTCTYVG
MAV_2872|M.avium_104 AETAFDRARKSLFEEGISTSRMGLDPARGGVEDLIDHITSGVRSTCTYVG
MMAR_2717|M.marinum_M TDSAFDRARKSLFEEGISTSRMGLDPDRGGVEDLIDHITSGVRSTCTYVG
MUL_3036|M.ulcerans_Agy99 TDSAFDRARKSLFEEGISTSRMGLDLDRGGVEDLIDHITSGVRSTCTYVG
MSMEG_3634|M.smegmatis_MC2_155 GDSSFDRARKGLFEEGISTSRMSLDPARGGVEDLLDHITSGVRSTCTYVG
TH_2384|M.thermoresistible__bu ADSPFDRARKALFEEGISTSRMGLDPSRGGVEDLLDHITAGLRSTCTYVG
MAB_2412c|M.abscessus_ATCC_199 GDSAFDQARKSLFEEGISTSRIELDPQRPGVEDLIDHIISGVRSTCTYVG
:..**:***.**:****:**: ** : *****:*** :*:********
Mflv_3360|M.gilvum_PYR-GCK AATLPELHDKAVLGVQSAAGFAEGHPLPTGW
Mvan_3090|M.vanbaalenii_PYR-1 ATTLSELHEKAVVGVQSAAGFAEGHPLPTGW
Mb1874c|M.bovis_AF2122/97 ASNLAELHERAVVGVQSGAGFAEGHPLPAGW
Rv1843c|M.tuberculosis_H37Rv ASNLAELHERAVVGVQSGAGFAEGHPLPAGW
MLBr_02066|M.leprae_Br4923 ASTLAELHEKAVVGVQSAAGFAEGHPLPSGW
MAV_2872|M.avium_104 AATLAQLYERVVVGVQSTAGFAEGHPLPLGW
MMAR_2717|M.marinum_M ASNLAELHEQVVIGVQSAAGFAEGHPLPAGW
MUL_3036|M.ulcerans_Agy99 ASNLAELHEQVVIGVQSAAGFAEGHPLPAGW
MSMEG_3634|M.smegmatis_MC2_155 AANLPELHEKVVLGVQSAAGFAEGHPLPAGW
TH_2384|M.thermoresistible__bu AATLPELHEKAVLGVQSQAGFAEGHPLPEGW
MAB_2412c|M.abscessus_ATCC_199 ARTLGELHERVVLGVQSAAGFAEGRPLPEGW
* .* :*:::.*:**** ******:*** **