For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPDSGIAALTPVTGLNVTLTDRVLSVRINRPSSLNSLTVPILTGIADTLERAAADPVVKVVRLGGVGRGF SSGVSMSVDDVWGGGPPTAIVEEANRAVRAVAALPHPVVAVVQGPAVGVAVSLALACDFILASDSAFFML ANTKVALMPDGGASALVAAATGRIRAMRLALLAEQLPAREALAWGLISAVYPDSDFEAEVDKVISRLLAG PALAFAQAKNAINAAALTELEPTFARELDGQEVLLRTHDFAEGAAAFLQRRTPNFTGS
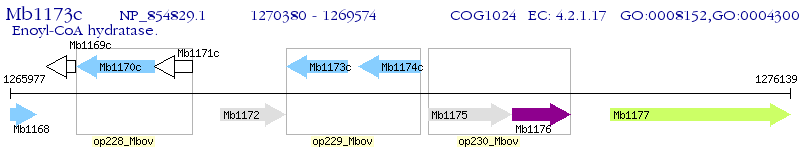
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1173c | echA11 | - | 100% (268) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb1174c | echA10 | e-103 | 72.28% (267) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb1099c | echA8 | 2e-23 | 29.27% (246) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2125 | - | 1e-74 | 58.04% (255) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv1141c | echA11 | 1e-146 | 100.00% (268) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-22 | 28.98% (245) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_4570c | - | 6e-43 | 38.37% (258) | enoyl-CoA hydratase/isomerase |
| M. marinum M | MMAR_4309 | echA10 | 5e-93 | 66.42% (268) | enoyl-CoA hydratase EchA10 |
| M. avium 104 | MAV_1283 | - | 1e-104 | 71.64% (268) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_5185 | - | 2e-80 | 61.33% (256) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_2638 | - | 3e-75 | 57.92% (259) | PUTATIVE enoyl-CoA hydratase PaaB |
| M. ulcerans Agy99 | MUL_0985 | echA10 | 2e-93 | 66.42% (268) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_4579 | - | 3e-83 | 61.54% (260) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1173c|M.bovis_AF2122/97 -----MPDSG------IAALTPVTGLNVTLTDRVLSVRINRPSSLNSLTV
Rv1141c|M.tuberculosis_H37Rv -----MPDSG------IAALTPVTGLNVTLTDRVLSVRINRPSSLNSLTV
MMAR_4309|M.marinum_M -----MPDTR------IETLAPVAGLEVKLRGGVLSVTIDRPESLNSLTP
MUL_0985|M.ulcerans_Agy99 -----MPDTR------IETLAPVAGLEVKLRGGVLSVTIDRPESLNSLTP
MAV_1283|M.avium_104 -----MPSSA------IATLAPVAGLDVTLSDGVFSVTINRPDSLNSLTV
MSMEG_5185|M.smegmatis_MC2_155 -----MTSTDASAETVTRDYPSIDDVSVSLTDGVLAVTLNRPDSLNSLNR
TH_2638|M.thermoresistible__bu -----MSS-------VT--YPSIDDLRVALSDGVLTVTLNRPDSLNSLTA
Mflv_2125|M.gilvum_PYR-GCK -----MTATD--------AYQGIDHLTASLDGSVLTVTLNRPESLNSLTA
Mvan_4579|M.vanbaalenii_PYR-1 MKGTFMTTTDP----TTDAYEGIADLAVGLDDGVLTVTLNRPNSLNSLTA
MAB_4570c|M.abscessus_ATCC_199 -----MSLDE-----------SVGAPDVAVDGGIMRVTFTRPDRMNALNG
MLBr_02402|M.leprae_Br4923 --------------------MKYDTILVDGYQRVGIITLNRPQALNALNS
. : : : **. :*:*.
Mb1173c|M.bovis_AF2122/97 PILTGIADTLERAAADPVVKVVRLGG--VGRGFSSGVSMSVDDVW-GGGP
Rv1141c|M.tuberculosis_H37Rv PILTGIADTLERAAADPVVKVVRLGG--VGRGFSSGVSMSVDDVW-GGGP
MMAR_4309|M.marinum_M EVMAGIADAMEAAATDSRVRVVRLGG--TGRGFSSGAGMSAQDMG-RKTS
MUL_0985|M.ulcerans_Agy99 EVMAGIADAMEAAATDSRVRVVRLGG--TGRGFSSGAGMSAQDMG-RKTS
MAV_1283|M.avium_104 PVITGIADAMEYAATDPEVKVVRIGG--AGRGFSSGAGISADDVS-DGGG
MSMEG_5185|M.smegmatis_MC2_155 DMLDAVADAMEVAATDPAVRVVRLGG--AGRGFSSGAGISEEDQN-AKG-
TH_2638|M.thermoresistible__bu PMLQTLAEAVELAASDPRVKVIRLTG--AGRGFCAGAGISEDDQA-ARES
Mflv_2125|M.gilvum_PYR-GCK PMLETFAVTLERAATDPRVRVVRIGG--AGRGFCSGAGISEEDHANPGAA
Mvan_4579|M.vanbaalenii_PYR-1 PMLQAFAATLERAAGDPRVRVVRIGG--AGRGFCSGAGISEEDHANPGAA
MAB_4570c|M.abscessus_ATCC_199 PATSGLIEALESIPGRDDVRVVVISGGAPGSAFSAGADVTELAAGSADLT
MLBr_02402|M.leprae_Br4923 QMMNEITNAAKELDIDPDVGAILITG--SPKVFAAGADIKEMASLTFTDA
. : : * .: : * *.:*..:.
Mb1173c|M.bovis_AF2122/97 --PTA--IVEEANRAVRAVAALPHPVVAVVQGPAVGVAVSLALACDFILA
Rv1141c|M.tuberculosis_H37Rv --PTA--IVEEANRAVRAVAALPHPVVAVVQGPAVGVAVSLALACDFILA
MMAR_4309|M.marinum_M --TSS---LTEINRAVRAITALPHPVVAVVHGPAAGVGVSLALACDLVLA
MUL_0985|M.ulcerans_Agy99 --TSS---LTEINRAVRAITALPHPVVAVVHGPAAGVGVSLALACDLVLA
MAV_1283|M.avium_104 --VPPDEIILEINRLVRAIAALPHPVVAVVQGPAAGVGVSIALACDVVLA
MSMEG_5185|M.smegmatis_MC2_155 --HDAAAVLDAANRAVAAIVALPKPVVAVVHGPAAGVGVSLALACDIVLA
TH_2638|M.thermoresistible__bu --GGGNALLDGANRSVTAIVNCPKPVVAVVPGAAAGVGVSLALACDVVLA
Mflv_2125|M.gilvum_PYR-GCK --GSPNDVLDAANRCIRAIANLPQPAVAVVQGAAAGVGVSLALACDVVLA
Mvan_4579|M.vanbaalenii_PYR-1 --GPPTDVLDGANRCIRAIANLPQPSVAVVQGAAAGVGVSLALACDVVIA
MAB_4570c|M.abscessus_ATCC_199 PLQAAELTMDNAERLVRAVLNCPVPTIAEVTGVAAGIGASVGLACDLIYA
MLBr_02402|M.leprae_Br4923 -------FDADFFSAWGKLAAVRTPMIAAVAGYALGGGCELAMMCDLLIA
: * :* * * * * . .:.: **.: *
Mb1173c|M.bovis_AF2122/97 SDSAFFMLANTKVALMPDGGASALVAAATGRIRAMRLALLAEQLPAREAL
Rv1141c|M.tuberculosis_H37Rv SDSAFFMLANTKVALMPDGGASALVAAATGRIRAMRLALLAEQLPAREAL
MMAR_4309|M.marinum_M SESAYFLLAFTKIGLMPDGGASALIAAAIGRIRAMRMALLPERLPATEAL
MUL_0985|M.ulcerans_Agy99 SESAYFLLAFTKIGLMPDGGASALIAAVIGRIRAMRMALLPERLPATEAL
MAV_1283|M.avium_104 SENAFFMLAFTKIGLMPDGGASALVAAAVGRIRAMQMALLPERLPAAEAL
MSMEG_5185|M.smegmatis_MC2_155 SDQAFFLLAFTKIGLMPDGGASALVAAAIGRIRAMRLALLADRLTATDAY
TH_2638|M.thermoresistible__bu SEQAFFLLAFTKIGLMPDGGASALVAAAIGRIRAMRMALLAERISAAEAH
Mflv_2125|M.gilvum_PYR-GCK SEKSFFMLAFTKIGLMPDGAASALIAAAVGRIRAMRMALLAERITADEAY
Mvan_4579|M.vanbaalenii_PYR-1 SEKAFFMLAFTKIGLMPDGGASALIAAAVGRIRAMRMALLAERISAREAY
MAB_4570c|M.abscessus_ATCC_199 ADTAFFLLAFANIGLMPDGGSSALVSASIGRVKANAMALLAQPLSATDAF
MLBr_02402|M.leprae_Br4923 ADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDLILTGRTIDAAEAE
:: : * . ::.::*. ..* :: *: :* : * : * :*
Mb1173c|M.bovis_AF2122/97 AWGLISAVYPDSDFEAEVDKVISRLLAGPALAFAQAKNAINAAALTELEP
Rv1141c|M.tuberculosis_H37Rv AWGLISAVYPDSDFEAEVDKVISRLLAGPALAFAQAKNAINAAALTELEP
MMAR_4309|M.marinum_M SYGLVSAVYPVADFQNEVDAVISRLLSGPAVAFAKTKDSINAATLTELDR
MUL_0985|M.ulcerans_Agy99 SYGLVSAVYPVADFQNEVDAVISRLLSGPAVAFAKTKNSINAATLTELDR
MAV_1283|M.avium_104 SWGLVTAVYPADEFEAEVDKVIARLLSGPAVAFAKTKLAINAATLTELDP
MSMEG_5185|M.smegmatis_MC2_155 DWGLISAVYPADEFDAAVDKVLAKLRNGPAVALRKTKQAINEATLTELDD
TH_2638|M.thermoresistible__bu QWGLVTGVYPADEFDAAVDKVISTLTNGPAVALRETKQAINAATLGELPD
Mflv_2125|M.gilvum_PYR-GCK AWGLVSQVYPADSLEAEAASVIETLATGPAVALRLTKDAINGATLTEVEN
Mvan_4579|M.vanbaalenii_PYR-1 DWGLVSSVHPAEEFDAEVDKVIATLVAGPAVALRKTKDAINAATLTELEA
MAB_4570c|M.abscessus_ATCC_199 AAGLVNEALPGDQLRERVDKTARRLAHGSRRAMQLTKEAMNAVSLKLFDE
MLBr_02402|M.leprae_Br4923 RSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAKEAVNRSFESTLAE
**:. . .: . . : . * :* ::* .
Mb1173c|M.bovis_AF2122/97 TFARELDGQEVLLRTHDFAEGAAAFLQRRTPNFTGS
Rv1141c|M.tuberculosis_H37Rv TFARELDGQEVLLRTHDFAEGAAAFLQRRTPNFTGS
MMAR_4309|M.marinum_M TLDREFQGQSALLRSPDFAEGARAFQQRRTPTFTDS
MUL_0985|M.ulcerans_Agy99 TLDREFQGQSALLRSPDFAEGARAFQQRRTPTFTDS
MAV_1283|M.avium_104 ALQREFDGQSVLLKSPDFVEGATAFQQRRTPNFTDR
MSMEG_5185|M.smegmatis_MC2_155 AIAREREGQLELLVANDFREGAQAFQQNRRPEFTDQ
TH_2638|M.thermoresistible__bu AIKRETEGQVRLLGSNDFREGTQAFQQKRAPKFTDV
Mflv_2125|M.gilvum_PYR-GCK ALERETKGQLRLLHSDDFREGTKAFQQRRAAKFTDV
Mvan_4579|M.vanbaalenii_PYR-1 ALERETAGQLVLLKSADFREGTHAFQQRRVAKFTDS
MAB_4570c|M.abscessus_ATCC_199 AIARERDGQIELLISPDAHEGFAAFLEGRRPNFS--
MLBr_02402|M.leprae_Br4923 GLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR
: :* : : * ** ** : * . *: