For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKELSVAEQRYQAVLAVISDGLSISQVAEKVGVSRQTLHTWLARYEAEGLDGLRIGTGTAL
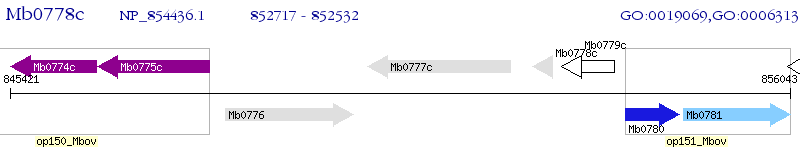
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0778c | - | - | 100% (61) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_4106 | - | 1e-09 | 53.06% (49) | transposase for ISMyma05 |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0573 | - | 3e-09 | 56.25% (48) | integrase catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0778c|M.bovis_AF2122/97 -MKELS------VAEQRYQAVLAVISDGLSISQVAEKVGVSRQTLHTWLA
Mvan_0573|M.vanbaalenii_PYR-1 MTQQLPNSPNDWVIEHRYRAVRQVL-DGVSKSQVAQECGASRQSVHSWVI
MMAR_4106|M.marinum_M MLQELR------MSEMRYRAVLEVL-DGAPVTAVARRFGVSRQTVHAWLR
::* : * **:** *: ** . : **.. *.***::*:*:
Mb0778c|M.bovis_AF2122/97 RYEAEG-LDGL-----------------------------------RIGT
Mvan_0573|M.vanbaalenii_PYR-1 RYEALG-VAGLADRSRRPLTSPNELSPAVVAMVCELRRTYPRWGAQRIAH
MMAR_4106|M.marinum_M RYAEEGAALNLEDRSSRPHRCPHQMPVEVEARVLTLRDAHPRWGPTRIVY
** * .* **
Mb0778c|M.bovis_AF2122/97 GTAL----------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 ELALRGVDAPPSRSSVYRILVRHGLVAAQQQNHKR-KYRRWQRDAPMQLW
MMAR_4106|M.marinum_M ELQRDVVPVVPGRSSVYRALVRNGRIDPAKRRRRRSDYKRWERGRPMELW
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 QIDIMGGVFLVDGRECKVVTGIDDHSRFVVMATVVADPGARAVCAAFTAT
MMAR_4106|M.marinum_M QMDVVGGLHLRDGIEVKVVTGIDDNSRFVVSAKVVARATARPVCAALLEA
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 MAIYGVPSEVLTDNGKQFTGRFTKP-YPAEVLFERICRENGITTRLTKPR
MMAR_4106|M.marinum_M LRRHGVPEQILTDNGKVFTGRFGPGGSSAEVLFDRVCVENGIGHLLTAPR
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 SPTTTGKIERFHKTLRRELLDSAG-PFASIEVAQEAIDAWVHGYNHSRPH
MMAR_4106|M.marinum_M SPTTTGKVERLHKTMRAEIFAEVDGVFDAIAELQAAIDRWVQYYNTARPH
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 QSLGMATPATMFRPAP----IEVVPPVPVDTEPLTPDVVVPPRPRRHLPP
MMAR_4106|M.marinum_M QSLGMVAPAARFALAAGPDLSVVEPVAAVPAEGQHPGALVDLRDAGVRRW
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 ATEHALIEVVHAVEWEAVLTPRARLLLPGDQQFKFTAALARREVTVWASD
MMAR_4106|M.marinum_M VNRHGSISVAG-------------------FRYRVPIVLAGEPVSVVVAD
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 RSIHIVLDGTVIRTRPSRLSEHDLRDLLRRGARIAGPEPARGAATIDILT
MMAR_4106|M.marinum_M NLVSIYHHDVLVASHVQHRKTDNTTGVVRQGRRTARPVTSG---------
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 TSAVIEVARTVSRDGCIGLGGDKVLLESTLVGQRVTLRFEGALMHVVANG
MMAR_4106|M.marinum_M ----IVVTRVVDSNGDVSFAGTMYRAGRVWRGKQVEVSIVAESVQIACAG
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 RLVKTMPAPLPPDQRATLRGARPTSEPLPPPAPPNRAMRRVAANGTITIA
MMAR_4106|M.marinum_M AIVRTHAIR---HDRAKEHGAFATPHGRP------RKPRQDAPDGPGAKA
Mb0778c|M.bovis_AF2122/97 --------------------------------------------------
Mvan_0573|M.vanbaalenii_PYR-1 GQRLRVGRSYQGRTVAVAIDDTVFRVFIDGTEICTHARRTDTDITIFKAY
MMAR_4106|M.marinum_M TP-------LRGRPEGRALTPAP---------------------------
Mb0778c|M.bovis_AF2122/97 ------
Mvan_0573|M.vanbaalenii_PYR-1 PRRHGT
MMAR_4106|M.marinum_M ------