For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MFVLNDDERVIVETAAAFAGKRLAPHALEWDAAKHFPVDVLREAAELGMAAIYCRDDVGGSGLRRLDGAR IFEQLAIADPVTAAFLSIHNMCAWMIDSFGTDEQRKDWIPRLATMGVIASYCLTEPGAGSDAGALSTRAV RHGSGKGGDYVLDGVKQFISGAAASDVYVVMARTGAEGPRGVSAFVVEKGTPGLSFGAPEAKMGWHAQPT AQVVLDGVRVPAEAMLGGADGEGAGFGIAMSGLNGGRLNIAACSLGGAQAAFDKAGAYVRDRQAFGGSLL DEPTVRFTLADMATGLQTSRMLLWRAASALDDDDADKVELCAMAKRYVTDTCFEVADQALQLHGGYGYLR EYGLEKIVRDLRVHRILEGTNEIMRLVIGRAEAARFRATV
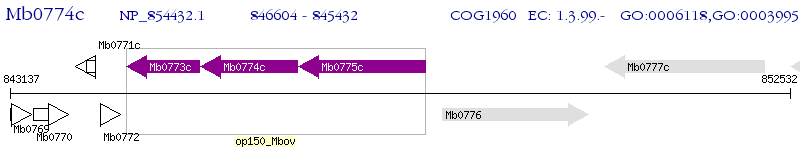
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0774c | fadE9 | - | 100% (390) | acyl-CoA dehydrogenase FADE9 |
| M. bovis AF2122 / 97 | Mb3302c | fadE25 | 3e-57 | 33.25% (382) | acyl-CoA dehydrogenase FADE25 |
| M. bovis AF2122 / 97 | Mb0221c | fadE3 | 2e-55 | 34.81% (385) | acyl-CoA dehydrogenase FADE3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5011 | - | 0.0 | 79.12% (388) | acyl-CoA dehydrogenase domain-containing protein |
| M. tuberculosis H37Rv | Rv0752c | fadE9 | 0.0 | 99.49% (390) | acyl-CoA dehydrogenase FADE9 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 2e-59 | 33.77% (382) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1188c | - | 1e-132 | 61.32% (380) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_1078 | fadE9 | 0.0 | 89.72% (389) | acyl-CoA dehydrogenase, FadE9 |
| M. avium 104 | MAV_4418 | - | 0.0 | 88.43% (389) | acyl-CoA dehydrogenase family protein member 8 |
| M. smegmatis MC2 155 | MSMEG_1497 | - | 1e-174 | 77.22% (395) | acyl-CoA dehydrogenase family protein member 8 |
| M. thermoresistible (build 8) | TH_4417 | - | 1e-179 | 79.17% (384) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_0836 | fadE9 | 0.0 | 89.20% (389) | acyl-CoA dehydrogenase, FadE9 |
| M. vanbaalenii PYR-1 | Mvan_1361 | - | 0.0 | 79.90% (388) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0774c|M.bovis_AF2122/97 -----------MFVLNDDERVIVETAAAFAGKRLAPHALEWDAAKHFPVD
Rv0752c|M.tuberculosis_H37Rv -----------MFVLNDDERVIVETAAAFAGKRLAPHALEWDAAKHFPVD
MMAR_1078|M.marinum_M -----------MFTLNDDERVITQTAAAFADKRLAPYALEWDATKHFPVD
MUL_0836|M.ulcerans_Agy99 -----------MFTLNDDERVITQTAAAFADKRLAPYALEWDATKHFPVD
MAV_4418|M.avium_104 -----------MFTLDDDERVIAETAAAFAAKRLAPHALEWDAAEHFPVD
Mflv_5011|M.gilvum_PYR-GCK ---------MDYFGLDDDDRVIAETAAAFAEKRLAPHALEWDETHHFPVD
Mvan_1361|M.vanbaalenii_PYR-1 ---------MDYFGLDEDERVIAETAAAFAEKRLAPYALEWDETHHFPAD
TH_4417|M.thermoresistible__bu ---------MDYLGLDDDERVIAETAAAFAEKRLAPYALEWDATKHFPTD
MSMEG_1497|M.smegmatis_MC2_155 ---------MYLSDLDDDERVIVETAAAFAEKRITPYALEWDDKHHFPTD
MAB_1188c|M.abscessus_ATCC_199 -----------MFNLTEEQREIWNTAREFADIHIAPHALDWDRDKYFPVE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
* ::.. : * :* .::*:* : * ** :
Mb0774c|M.bovis_AF2122/97 VLREAAELGMAAIYCRDDVGGSGLRRLDGARIFEQLAIADPVTAAFLSIH
Rv0752c|M.tuberculosis_H37Rv VLREAAELGMAAIYCRDDVGGSGLRRLDGVRIFEQLAIADPVTAAFLSIH
MMAR_1078|M.marinum_M VLREAAELGMAAIYCREDVGGSGLRRLDGVRIFEQLAIADPVTAAFLSIH
MUL_0836|M.ulcerans_Agy99 VLREAAELGMAAIYCREDVGGSGLRRLDGVRIFEQLAIADPVTAAFLSIH
MAV_4418|M.avium_104 VLRESAELGMAAIYCREDVGGSGLRRLDGVRIFEQLAIADPTTAAFLSIH
Mflv_5011|M.gilvum_PYR-GCK VLREAAELGMAAIYCGEDAGGSGLRRIDAVRIFEKLSTADPTVAAFLSIH
Mvan_1361|M.vanbaalenii_PYR-1 VLREAAELGMAAIYCRDDVGGSGLRRIDAVRIFEKLATADPTVAAFLSIH
TH_4417|M.thermoresistible__bu ALREAAELGMAAIYCSEEVGGSGLRRLDAVRIFEHLSAADPTTAAFLSIH
MSMEG_1497|M.smegmatis_MC2_155 VLREAAELGMGAIYCREDVGGSELRRLDAVRIFEALSAADPTLAAFLSIH
MAB_1188c|M.abscessus_ATCC_199 IFPKAAALGMGGIYVNPDVGGSGLTRLDASLIFEAMATGCSAVSAFISIH
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
: *: .*: : **. : . ::* :: . : : :::
Mb0774c|M.bovis_AF2122/97 NMCAWMIDSFGTDEQRKDWIPRLATMGVIASYCLTEPGAGSDAGALSTRA
Rv0752c|M.tuberculosis_H37Rv NMCAWMIDSFGTDEQRKDWIPRLATMGVIASYCLTEPGAGSDAGALSTRA
MMAR_1078|M.marinum_M NMCAWMIDSFGTDEQRKDWIPRLATMDAIASYCLTEPGAGSDASALSARA
MUL_0836|M.ulcerans_Agy99 NMCTWMIDSFGTDEQRKDWIPRLATMDAIASYCLTEPGAGSDASALSARA
MAV_4418|M.avium_104 NMCAWMIDSYGTPEQRKGWVPRLAAMDVIASYCLTEPGAGSDAAALSTRA
Mflv_5011|M.gilvum_PYR-GCK NMCAWMVDSFGTEEQRKLWVPKLASMDAIASYCLTEPGAGSDASALRTKA
Mvan_1361|M.vanbaalenii_PYR-1 NMCAWMVDSFGTEEQRKEWVPRLASMDAIASYCLTEPGAGSDASALRTKA
TH_4417|M.thermoresistible__bu NMCAWMVDTYGTDEQRKTWVPRLASMDAIASYCLTEPGAGSDAAALRTRA
MSMEG_1497|M.smegmatis_MC2_155 NMCAWMVDTYGTEDQRKTWVPRLASMETIASYCLTEPGAGSDAAALRTRA
MAB_1188c|M.abscessus_ATCC_199 NMAAWMIDEFGDEEQRRRWLPSMCTMETIGAYCLTEPECGSDAAALRTSA
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
:: : : * : :: :* :.: .:.:*.*:* .****.:: : *
Mb0774c|M.bovis_AF2122/97 VRHGSGKGGDYVLDGVKQFISGAAASDVYVVMARTGAE-GPRGVSAFVVE
Rv0752c|M.tuberculosis_H37Rv VRHGSGKGGDYVLDGVKQFISGAAASDVYVVMARTGAE-GPRGVSAFIVE
MMAR_1078|M.marinum_M VRQGSAPGSDYVLDGVKQFISGAGASDVYVVMARTGAE-GPRGISAFIVE
MUL_0836|M.ulcerans_Agy99 VRQGSAPGSDYVLDGVKQFISGAGASDVYVVMARTGAE-GPRGISAFIVE
MAV_4418|M.avium_104 VRQ----GGDYVLDGVKQFISGAGASDVYLVMARTGGE-GPRGISAFVVE
Mflv_5011|M.gilvum_PYR-GCK VRS----GDEYVLDGVKQFISGAGTSDVYIVMARTGSD-GPKGISAFVVP
Mvan_1361|M.vanbaalenii_PYR-1 VRS----GDDYVLDGVKQFISGAGSSDVYVVMARTGSD-GPKGISAFVVP
TH_4417|M.thermoresistible__bu VRD----GDHFVLDGVKQFISGAGASDVYVVMARTGED-GPKGISTFIVE
MSMEG_1497|M.smegmatis_MC2_155 VKD----GDHYVLDGVKQFISGAGTSDLYVVMARTGGE-GPRGISTFIVE
MAB_1188c|M.abscessus_ATCC_199 IRQ----GDEYVLNGVKQFISGAGAADLYVVMARTGGP-GPRGISTIVVP
MLBr_00737|M.leprae_Br4923 KAD----GDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGANGISAFIVH
*..::*:*.* :*:... : * *** *. *..*:*:::*
Mb0774c|M.bovis_AF2122/97 KGTPGLSFGAPEAKMGWHAQPTAQVVLDGVRVPAEAMLGGADGEGAGFGI
Rv0752c|M.tuberculosis_H37Rv KGTPGLSFGAPEAKMGWHAQPTAQVVLDGVRVPAEAMLGGADGEGAGFGI
MMAR_1078|M.marinum_M KGTPGLSFGALEEKMGWHAQPTAQVVLEGVRVPADAMLGGVDGTGAGFGI
MUL_0836|M.ulcerans_Agy99 KGTPGLSFGALEEKMGWHAQPTAQVVLEGVRVPADAMLGGVDGTGAGFGI
MAV_4418|M.avium_104 KGSAGLSFGSPEEKMGWHAQPTAQVILEGVRVPADAMLGGAEGEGAGFGI
Mflv_5011|M.gilvum_PYR-GCK KDTPGLSFGTDEVKMGWNAQPTRQVILEGARVPADALLGGPDGEGTGFGI
Mvan_1361|M.vanbaalenii_PYR-1 KDTPGLSFGTDEQKMGWNAQPTKQVIFEGARVPADALLGGPDGEGTGFGI
TH_4417|M.thermoresistible__bu KGTPGLSFGADEQKMGWNAQPTAQVILEGARVPAENMLGGPEGEGKGFSI
MSMEG_1497|M.smegmatis_MC2_155 KDTPGLSFGADEQKMGWNAQPTAQVIFEGVRVPAEALLG--NTEGTGFGI
MAB_1188c|M.abscessus_ATCC_199 KGTPGLSFGAPERKMGWHAQPTAQVVFEDVRVPVANRIG---EEGIGFTI
MLBr_00737|M.leprae_Br4923 KDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIG---EPGTGFKT
*. *:*:*. * *:* :..** :: :: *:* :* * **
Mb0774c|M.bovis_AF2122/97 AMSGLNGGRLNIAACSLGGAQAAFDKAGAYVRDRQAFGGSLLDEPTVRFT
Rv0752c|M.tuberculosis_H37Rv AMSGLNGGRLNIAACSLGGAQAAFDKAGAYVRDRQAFGGSLLDEPTVRFT
MMAR_1078|M.marinum_M AMNGLNGGRLNIAACSLGGAQSAFAKAGAYVRERQAFGASLLDEPTVRFT
MUL_0836|M.ulcerans_Agy99 AMNGLNGGRLNIAACSLGGAQSAFAKTGAYVRERQAFGASLLDEPTVRFT
MAV_4418|M.avium_104 AMNGLNGGRLNIAACSLGGAQAAFDKAGAYVRDRQAFGSPLLDEPTIRFA
Mflv_5011|M.gilvum_PYR-GCK AMKGLNGGRINIAACSLGGAQAAYDKTVSYLADRQAFGGSLLDEPTIRFT
Mvan_1361|M.vanbaalenii_PYR-1 AMKGLNGGRINIAACSLGGAQTAYDKSAAYLADRQAFGGALLDEPTIRFT
TH_4417|M.thermoresistible__bu AMNGLNGGRLNIAACSLGGAQTAYEKAASYVADRQAFGGSLIDEPTIRFT
MSMEG_1497|M.smegmatis_MC2_155 AMNGLNGGRINIAACSLGGAQAAYDKAVGYLADRQAFGAALLDEPTIRFT
MAB_1188c|M.abscessus_ATCC_199 AMRGLNGGRLNIASCSLGGARSALEKVIEYLRTRKAFGEELIKFQALQFR
MLBr_00737|M.leprae_Br4923 ALATLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFM
*: *: * .*.: ::* *: * * *: ** : :::*
Mb0774c|M.bovis_AF2122/97 LADMATGLQTSRMLLWRAASALDDDDADKVELCAMAKRYVTDTCFEVADQ
Rv0752c|M.tuberculosis_H37Rv LADMATGLQTSRMLLWRAASALDDDDADKVELCAMAKRYVTDTCFEVADQ
MMAR_1078|M.marinum_M LADMATGLEASRMLLWRAANALDEDDADKVELCAMAKRFVTDTCFDVADK
MUL_0836|M.ulcerans_Agy99 LADMATGLEASRMLLWRAANALDEDDADKVELCAMAKRFVTDTCFDVADK
MAV_4418|M.avium_104 LADMATGLETSRMMLWRAAGALDADDPDKVELCAMAKRYVTDTCFEVADK
Mflv_5011|M.gilvum_PYR-GCK LADMATGLETSRALLWRAATALDNDHPDRVELCAMAKRYVTDACFEVADQ
Mvan_1361|M.vanbaalenii_PYR-1 LADMATALETSRVLLWRAATALDNDHPDRVELCAMAKRYVTDACFEVADQ
TH_4417|M.thermoresistible__bu LADMATALQTSRLMLWRAATALDENHPDKVELCAMAKLHVTEACYEVADK
MSMEG_1497|M.smegmatis_MC2_155 LADMATGLETSRNLLWRAASALDANHPDKVTLCAMAKRYVTDTCFDVADS
MAB_1188c|M.abscessus_ATCC_199 LADMATELEAARTMVWRAAAAVTEGDPRAAELCAMAKRIATDVGFTVANE
MLBr_00737|M.leprae_Br4923 LADMAMKVEAARLIVYAAAARAERGEPDLGFISAASKCFASDIAMEVTTD
***** ::::* ::: ** ... :.* :* .:: *: .
Mb0774c|M.bovis_AF2122/97 ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRLVIGRAEAARFRA
Rv0752c|M.tuberculosis_H37Rv ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRLVIGRAEAARFRA
MMAR_1078|M.marinum_M ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRVVVGRAEAARFRA
MUL_0836|M.ulcerans_Agy99 ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRVVVGRAEAARFRA
MAV_4418|M.avium_104 ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRVVIGRAEAARFRA
Mflv_5011|M.gilvum_PYR-GCK ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRVVIGRSAASRARA
Mvan_1361|M.vanbaalenii_PYR-1 ALQLHGGYGYLREYGLEKIVRDLRVHRILEGSNEIMRVVIGRAAAARARA
TH_4417|M.thermoresistible__bu ALQLHGGYGYLNEYGLEKIVRDLRVHRILEGTNEIMRVVIGRAVAGRARG
MSMEG_1497|M.smegmatis_MC2_155 ALQLHGGYGYLREYGLEKIVRDLRVHRILEGTNEIMRVVIGRSVAGAARN
MAB_1188c|M.abscessus_ATCC_199 ALQLHGGYGYLAEYGIEKIVRDLRVHQILEGTNEIMRVIVSRKLIEQGLP
MLBr_00737|M.leprae_Br4923 AVQLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR----
*:**.** ** :: :*:::** :: :* **:*:* *:::.*
Mb0774c|M.bovis_AF2122/97 TV---------
Rv0752c|M.tuberculosis_H37Rv TV---------
MMAR_1078|M.marinum_M SA---------
MUL_0836|M.ulcerans_Agy99 SA---------
MAV_4418|M.avium_104 TA---------
Mflv_5011|M.gilvum_PYR-GCK S----------
Mvan_1361|M.vanbaalenii_PYR-1 S----------
TH_4417|M.thermoresistible__bu AAESGRARASA
MSMEG_1497|M.smegmatis_MC2_155 QADAARPRATA
MAB_1188c|M.abscessus_ATCC_199 A----------
MLBr_00737|M.leprae_Br4923 -----------