For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVDAHRGGHPTPMSSTKATLRLAEATDSSGKITKRGADKLISTIDEFAKIAISSGCAELMAFATSAVRDA ENSEDVLSRVRKETGVELQALRGEDESRLTFLAVRRWYGWSAGRILNLDIGGGSLEVSSGVDEEPEIALS LPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELAEPSVTVLEAGSPDLAVATSKTFRSLARLTGAAPSM AGPRVKRTLTANGLRQLIAFISRMTAVDRAELEGVSADRAPQIVAGALVAEASMRALSIEAVEICPWALR EGLILRKLDSEADGTALIESSSVHTSVRAVGGQPADRNAANRSRGSKP
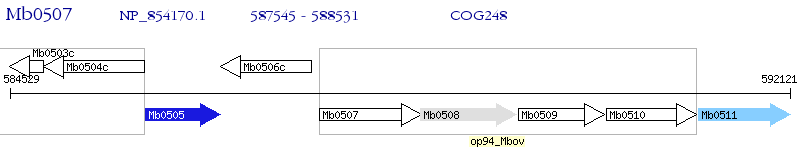
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0507 | - | - | 100% (328) | hypothetical protein Mb0507 |
| M. bovis AF2122 / 97 | Mb1054 | - | 6e-13 | 25.88% (255) | hypothetical protein Mb1054 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0079 | - | 1e-142 | 79.69% (320) | Ppx/GppA phosphatase |
| M. tuberculosis H37Rv | Rv0496 | - | 0.0 | 100.00% (328) | hypothetical protein Rv0496 |
| M. leprae Br4923 | MLBr_02434 | - | 1e-157 | 85.93% (327) | hypothetical protein MLBr_02434 |
| M. abscessus ATCC 19977 | MAB_4013c | - | 1e-138 | 82.27% (299) | hypothetical protein MAB_4013c |
| M. marinum M | MMAR_0822 | - | 1e-160 | 87.20% (328) | hypothetical protein MMAR_0822 |
| M. avium 104 | MAV_4656 | - | 1e-156 | 86.89% (328) | Ppx/GppA phosphatase family protein |
| M. smegmatis MC2 155 | MSMEG_0939 | - | 1e-145 | 85.10% (302) | Ppx/GppA phosphatase family protein |
| M. thermoresistible (build 8) | TH_0881 | - | 1e-139 | 83.33% (294) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_4566 | - | 1e-159 | 86.89% (328) | hypothetical protein MUL_4566 |
| M. vanbaalenii PYR-1 | Mvan_0833 | - | 1e-143 | 84.23% (298) | Ppx/GppA phosphatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0079|M.gilvum_PYR-GCK MRLGVLDVGSNTVHLLVVDARRGGHPTPMSSTKASLRLAEAIDDSGKLTR
Mvan_0833|M.vanbaalenii_PYR-1 MRLGVLDVGSNTVHLLVVDARRGGHPTPMSATKASLRLAESIDNSGKLTR
Mb0507|M.bovis_AF2122/97 ----------------MVDAHRGGHPTPMSSTKATLRLAEATDSSGKITK
Rv0496|M.tuberculosis_H37Rv ----------------MVDAHRGGHPTPMSSTKATLRLAEATDSSGKITK
MMAR_0822|M.marinum_M MRLGVLDVGSNTVHLLVVDAHRGGHPTPMSSTKATLRLAEATDSSGNITK
MUL_4566|M.ulcerans_Agy99 MRLGVLDVGSNTVHLLVVDAHRGGHPTPMSSTKATLRLAEATDSSGNITK
MLBr_02434|M.leprae_Br4923 MRLGVLDVGSNTVHLLVVDAYRGGHPTPMSSTKATLRMVEATDSSGKITK
MAV_4656|M.avium_104 MRLGVLDVGSNTVHLLVVDAHRGGHPTPMSSTKATLRLAEATDSAGKITK
MAB_4013c|M.abscessus_ATCC_199 MRLGVLDVGSNTVHLLVVDARRGGHPTPMSSTKAALRLAEAIDESGKLTR
MSMEG_0939|M.smegmatis_MC2_155 ----MLDVGSNTVHLLVVDAHRGGHPTPMSSTKAALRLAEAIDGSGKLTR
TH_0881|M.thermoresistible__bu ----VLDVGSNTVHLLIVDARRGGHPTPMSSTKASLRLAEQIDGSGKITR
:*** *********:***:**:.* * :*::*:
Mflv_0079|M.gilvum_PYR-GCK RGADKLIGTLDEFARIATSSGCAELMAFATSAVRDARNSEEVLARVKAET
Mvan_0833|M.vanbaalenii_PYR-1 RGADKLVGTIDEFARIATSSGCAELMAFATSAVRDATNSEDVLARVRAET
Mb0507|M.bovis_AF2122/97 RGADKLISTIDEFAKIAISSGCAELMAFATSAVRDAENSEDVLSRVRKET
Rv0496|M.tuberculosis_H37Rv RGADKLISTIDEFAKIAISSGCAELMAFATSAVRDAENSEDVLSRVRKET
MMAR_0822|M.marinum_M RGADKLVSTIDEFAKIAASSGCAELMAFATSAVREAENSDDVLSRVRNET
MUL_4566|M.ulcerans_Agy99 RGADKLVSTIDEFAKIAASSGCAELMAFATSAVREAENSDDVLSRVRNET
MLBr_02434|M.leprae_Br4923 RAADKLVSTIGEFAKIAVSSGCAELMAFATSAVREAGNSDDVLSRVRKET
MAV_4656|M.avium_104 RGAEKLISTIDEFAKIADSSGCEELMAFATSAVREAGNSEEVLNRVRKET
MAB_4013c|M.abscessus_ATCC_199 KGADSLVHTIGEFSKIARSSGCTEIMAFATSAVRDATNSDEVLSRVKKET
MSMEG_0939|M.smegmatis_MC2_155 KGADKLVATVDEFAKIATSSGCAELMAFATSAVRDATNSEDVLARVRAET
TH_0881|M.thermoresistible__bu KGADKLISTIGEFARIAASSGCAELMAFATSAVRDATNTEEVLERVRAET
:.*:.*: *:.**::** **** *:*********:* *:::** **: **
Mflv_0079|M.gilvum_PYR-GCK GVELKVLSGVDESRLTFLAVRRWYGWSAGRIVNIDIGGGSLELSSGVDEE
Mvan_0833|M.vanbaalenii_PYR-1 GVDLRVLSGVDESRLTFLAVRRWYGWSAGRIINIDIGGGSLELSSGVDEE
Mb0507|M.bovis_AF2122/97 GVELQALRGEDESRLTFLAVRRWYGWSAGRILNLDIGGGSLEVSSGVDEE
Rv0496|M.tuberculosis_H37Rv GVELQALRGEDESRLTFLAVRRWYGWSAGRILNLDIGGGSLEVSSGVDEE
MMAR_0822|M.marinum_M DVELQVLSGVDESRLTFLAVRRWFGWSAGRIINLDIGGGSLELSSGVDEE
MUL_4566|M.ulcerans_Agy99 GVELQVLSGVDESRLTFLAVRRWFGWSAGRIINLDIGGGSLELSSGVDEE
MLBr_02434|M.leprae_Br4923 GVRLQVLRGVDESRLTFLAVRRWFGWSAGRIINLDIGGGSLELSNGVDEE
MAV_4656|M.avium_104 GVELRVLTGVDESRLTFLAVRRWYGWSAGRIINLDIGGGSLEMSSGLDEE
MAB_4013c|M.abscessus_ATCC_199 GVDLAVLSGVNESRLTFLAVRRWYGWSAGRIINLDIGGGSLEMSCGVDEE
MSMEG_0939|M.smegmatis_MC2_155 GVSLQVLSGVDESRLTFLAVRRWYGWSAGRIINIDIGGGSLELSNGVDEE
TH_0881|M.thermoresistible__bu GVSLQVLRGVDESRLTFLAVRRWYGWSAGRIINIDIGGGSLELSNGVDEE
.* * .* * :************:*******:*:********:* *:***
Mflv_0079|M.gilvum_PYR-GCK PDVALSLPLGAGRMTREWLPDDPPGRRRVAMLRDWLTNELSDAAAAVTEA
Mvan_0833|M.vanbaalenii_PYR-1 PDVALSLPLGAGRMTREWLPDDPPGRRRVAVLRDWLSNELSEPAAAIAEA
Mb0507|M.bovis_AF2122/97 PEIALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELAEPSVTVLEA
Rv0496|M.tuberculosis_H37Rv PEIALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELAEPSVTVLEA
MMAR_0822|M.marinum_M PEVALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELSDASVTMLDA
MUL_4566|M.ulcerans_Agy99 PGVALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELPDASVTMLDA
MLBr_02434|M.leprae_Br4923 PEVALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDSELSDASVTVLEA
MAV_4656|M.avium_104 PEVALSLPLGAGRLTREWLPDDPPGRRRVAMLRDWLDAELAEASENILEA
MAB_4013c|M.abscessus_ATCC_199 PDIALSLPLGAGRLTREWLAEDPPGRRRVAMLRDWLESELVDAAKQVAAA
MSMEG_0939|M.smegmatis_MC2_155 PEVALSLPLGAGRLTREWLPDDPPGRRAVAMLRDWLATELADAGATIRRA
TH_0881|M.thermoresistible__bu PDVALSLPLGAGRLSREWLPDDPPGRRRVAMLRDWLATELAEPAATVLAA
* :**********::****.:****** **:***** ** :.. : *
Mflv_0079|M.gilvum_PYR-GCK GTPDLAVASSKTFRSLARLTGAAPSGAGPRVKRTLTATGLRQLINFISRM
Mvan_0833|M.vanbaalenii_PYR-1 GAPDLAVASSKTFRSLARLTGAAPSGAGPRVKRTLTASGLRQLINFISRM
Mb0507|M.bovis_AF2122/97 GSPDLAVATSKTFRSLARLTGAAPSMAGPRVKRTLTANGLRQLIAFISRM
Rv0496|M.tuberculosis_H37Rv GSPDLAVATSKTFRSLARLTGAAPSMAGPRVKRTLTANGLRQLIAFISRM
MMAR_0822|M.marinum_M GIPDLAVATSKTFRSLARLTGAAPSAAGPRVKRTLTANGLRQLISFISRM
MUL_4566|M.ulcerans_Agy99 GIPDLAVATSKTFRSLARLTGAAPSAAGPRVKRTLTANGLRQLISFISRM
MLBr_02434|M.leprae_Br4923 GKPDLAVATSKTFRSLARLTGAAPSAAGPRAKRALTVNGLRQLIAFISRM
MAV_4656|M.avium_104 GTPDLTVATSKTFRSLARLTGAAPSGAGPRVKRTLTANGLRQLISFISRM
MAB_4013c|M.abscessus_ATCC_199 GVPDLAVASSKTFRSLARLTGAAPSGAGPRVKRTLTASGLRQLIAFISRM
MSMEG_0939|M.smegmatis_MC2_155 GAPDLAVATSKTFRSLARLTGAAPSGAGPRVKRTLTATGLRQLIAFISRM
TH_0881|M.thermoresistible__bu GSPDLVVASSKTFHSLARLTGAAPSAAGPRVRRTVTATGLRQVIAFISRM
* ***.**:****:*********** ****.:*::*..****:* *****
Mflv_0079|M.gilvum_PYR-GCK TAADRAELEGVSADRAPQIVAGALVAEASMNALNIESVDICPWALREGLI
Mvan_0833|M.vanbaalenii_PYR-1 TAADRAELEGVSADRAPQIVAGALVAEASMKALGIESVDICPWALREGLI
Mb0507|M.bovis_AF2122/97 TAVDRAELEGVSADRAPQIVAGALVAEASMRALSIEAVEICPWALREGLI
Rv0496|M.tuberculosis_H37Rv TAVDRAELEGVSADRAPQIVAGALVAEASMRALSIEAVEICPWALREGLI
MMAR_0822|M.marinum_M TTADRAELEGVSAERAPQIVAGALVAEASMRALSIEALDICPWALREGVI
MUL_4566|M.ulcerans_Agy99 TTADRAELEGVSAERARQIVAGALVAEASMRALSIEALDICPWALREGVI
MLBr_02434|M.leprae_Br4923 TASDRAELEGISTERAPQIVAGALVAEASMRALSIETVDICPWALREGLI
MAV_4656|M.avium_104 TTADRAELEGVSAERAPQIVAGALVAEASMRALSIESVDICPWALREGLI
MAB_4013c|M.abscessus_ATCC_199 TTADRAELEGVSVDRAPQIVAGALVAEASMRALSLEAVDICPWALREGLI
MSMEG_0939|M.smegmatis_MC2_155 TAADRAELEGVSAERAPQIVAGALVAEASMRALEIESVDICPWALREGLI
TH_0881|M.thermoresistible__bu TTADRAELEGVSADRAPQIVAGALIAEAAMKALEVETVDICPWALREGLI
*: *******:*.:** *******:***:*.** :*:::*********:*
Mflv_0079|M.gilvum_PYR-GCK LRKLDSEADGSD------FVGGSFN--------QVSGRGKAAKT-GARES
Mvan_0833|M.vanbaalenii_PYR-1 LRKLDSEADGSA------LVGGTDDSGGSGKPGKVGGTDKAGKADGGESA
Mb0507|M.bovis_AF2122/97 LRKLDSEADGTA------LIESSSV--------HTSVRAVGGQPADRNAA
Rv0496|M.tuberculosis_H37Rv LRKLDSEADGTA------LIESSSV--------HTSVRAVGGQPADRNAA
MMAR_0822|M.marinum_M LRRLDSEGDGTA------FMETSPV--------LTSVRDARGQVVDRNAA
MUL_4566|M.ulcerans_Agy99 LRRLDSEGDGTA------FMETSPV--------LTSVRDARGQVVDRNAA
MLBr_02434|M.leprae_Br4923 LRKLDSEADGTA------LVQTS-------------VRDTRGQEVDRNAA
MAV_4656|M.avium_104 LRKLDSEADGTA------LMEPS-------------VRNAGGQVVDRNQ-
MAB_4013c|M.abscessus_ATCC_199 LRKLDSEADGTA------FVD-----------------------------
MSMEG_0939|M.smegmatis_MC2_155 LRKLDSEADGTA------LVAAAAR-----------------------DA
TH_0881|M.thermoresistible__bu LRKLDSEAVGGSGTDHPFDVGGADNPATIDADQFATVSPSAREAGAMHPA
**:****. * :
Mflv_0079|M.gilvum_PYR-GCK ARTADART-----
Mvan_0833|M.vanbaalenii_PYR-1 ARPMSVRNAGR--
Mb0507|M.bovis_AF2122/97 NRSRGSKP-----
Rv0496|M.tuberculosis_H37Rv NRSRGSKP-----
MMAR_0822|M.marinum_M GRSRGNKP-----
MUL_4566|M.ulcerans_Agy99 GRSRGNKP-----
MLBr_02434|M.leprae_Br4923 NRSRGDKT-----
MAV_4656|M.avium_104 NRSRGDKP-----
MAB_4013c|M.abscessus_ATCC_199 -------------
MSMEG_0939|M.smegmatis_MC2_155 GR-----------
TH_0881|M.thermoresistible__bu STTSPAGPRGNTR